bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3510_orf3
Length=435
Score E
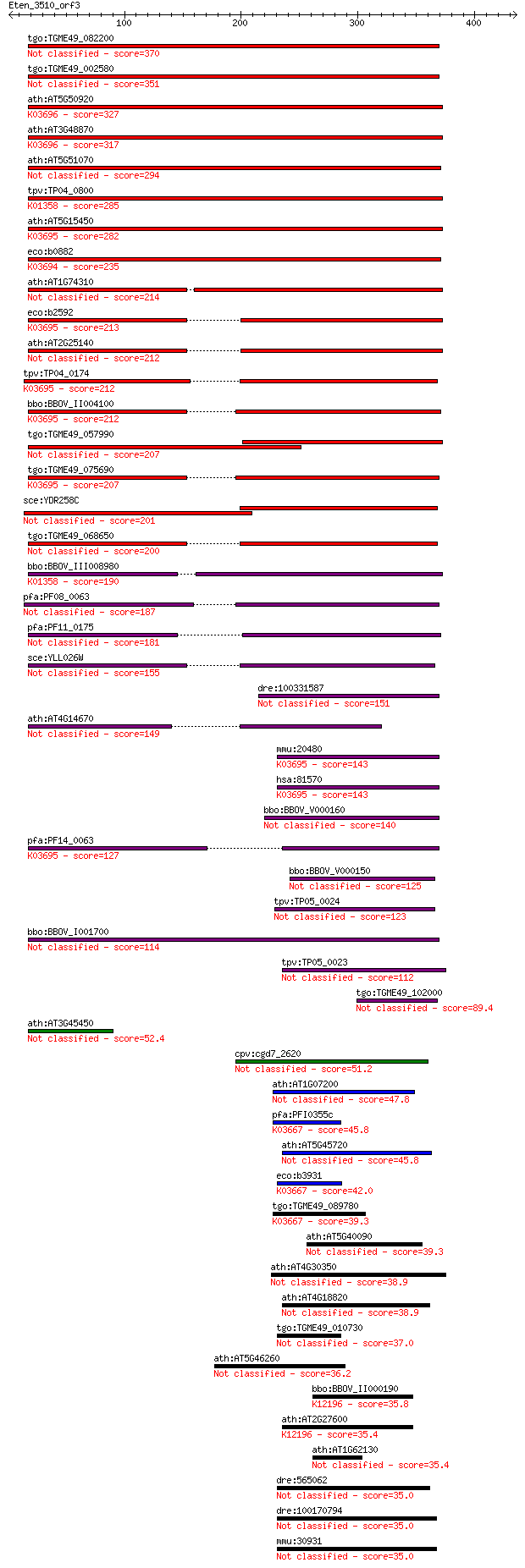
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_082200 clpB protein, putative 370 6e-102
tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53) 351 3e-96
ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptid... 327 6e-89
ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding / n... 317 5e-86
ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1); ... 294 4e-79
tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit (... 285 3e-76
ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP b... 282 2e-75
eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity ... 235 2e-61
ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEI... 214 4e-55
eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation ... 213 2e-54
ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP b... 212 2e-54
tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp p... 212 2e-54
bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp pr... 212 3e-54
tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53) 207 4e-53
tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.2... 207 8e-53
sce:YDR258C HSP78; Hsp78p 201 5e-51
tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.2... 200 8e-51
bbo:BBOV_III008980 17.m07783; Clp amino terminal domain contai... 190 8e-48
pfa:PF08_0063 ClpB protein, putative 187 9e-47
pfa:PF11_0175 heat shock protein 101, putative 181 6e-45
sce:YLL026W HSP104; Heat shock protein that cooperates with Yd... 155 3e-37
dre:100331587 suppressor of K+ transport defect 3-like 151 6e-36
ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosph... 149 2e-35
mmu:20480 Clpb, AL118244, Skd3; ClpB caseinolytic peptidase B ... 143 1e-33
hsa:81570 CLPB, FLJ13152, HSP78, SKD3; ClpB caseinolytic pepti... 143 1e-33
bbo:BBOV_V000160 clpC 140 7e-33
pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP... 127 8e-29
bbo:BBOV_V000150 clpC 125 2e-28
tpv:TP05_0024 clpC; molecular chaperone 123 1e-27
bbo:BBOV_I001700 19.m02115; chaperone clpB 114 6e-25
tpv:TP05_0023 clpC; molecular chaperone 112 4e-24
tgo:TGME49_102000 chaperone clpB protein, putative 89.4 2e-17
ath:AT3G45450 Clp amino terminal domain-containing protein 52.4 4e-06
cpv:cgd7_2620 ClpB ATpase (bacterial), signal peptide 51.2 7e-06
ath:AT1G07200 ATP-dependent Clp protease ClpB protein-related 47.8 8e-05
pfa:PFI0355c ATP-dependent heat shock protein, putative; K0366... 45.8 3e-04
ath:AT5G45720 ATP binding / DNA binding / DNA-directed DNA pol... 45.8 3e-04
eco:b3931 hslU, clpY, ECK3923, htpI, JW3902; molecular chapero... 42.0 0.005
tgo:TGME49_089780 ATP-dependent heat shock protein, putative (... 39.3 0.034
ath:AT5G40090 ATP binding / nucleoside-triphosphatase/ nucleot... 39.3 0.034
ath:AT4G30350 heat shock protein-related 38.9 0.036
ath:AT4G18820 ATP binding / DNA binding / DNA-directed DNA pol... 38.9 0.039
tgo:TGME49_010730 ATP-dependent protease ATP-binding subunit, ... 37.0 0.14
ath:AT5G46260 disease resistance protein (TIR-NBS-LRR class), ... 36.2 0.23
bbo:BBOV_II000190 18.m05996; ATPase, AAA family; K12196 vacuol... 35.8 0.35
ath:AT2G27600 SKD1; SKD1 (SUPPRESSOR OF K+ TRANSPORT GROWTH DE... 35.4 0.42
ath:AT1G62130 AAA-type ATPase family protein 35.4 0.50
dre:565062 im:7165310; si:ch73-178d14.1 35.0 0.63
dre:100170794 zgc:194342 35.0 0.63
mmu:30931 Tor1a, DQ2, Dyt1, MGC18883, torsinA; torsin family 1... 35.0 0.65
> tgo:TGME49_082200 clpB protein, putative
Length=970
Score = 370 bits (950), Expect = 6e-102, Method: Compositional matrix adjust.
Identities = 192/372 (51%), Positives = 260/372 (69%), Gaps = 26/372 (6%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DE+H L+GAGK+ G+MDA+ +LK P+ARGEI L+GATT EYK+ +EKDAA RR + I
Sbjct 354 DELHMLMGAGKSDGTMDAANLLKPPMARGEIRLVGATTQEEYKI-IEKDAAMERRLKPIF 412
Query 78 VEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDEA 137
+E PS +R + ILRK+ +E+HH ++I D + V LS + ++ R PDKA+DLLDEA
Sbjct 413 IEEPSTDRAIYILRKLSDKFESHHEMKISDEAIVAAVMLSHKYIRNRKLPDKAIDLLDEA 472
Query 138 CAFRRVRHNSRRA---ELALLLQQQRDGKTHFAEEELAALQAEYAEMTEPT--------- 185
A +RV+ + R E+A+ +R F EE Q + + +
Sbjct 473 AATKRVKWDLRSLNDDEVAM----ERKNSEQFEEETRKVEQQDQQRIEKQNHGDGTEEEL 528
Query 186 -------EERP--DPEVLELQMDDVAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKRVI 236
EE+ + + L L DDVA V+SLWTGIP+ ++T+DE +++L+L+DLL RVI
Sbjct 529 KNLEKEIEEQSLHEKQELVLTADDVAQVISLWTGIPLAKLTDDEKSKILRLSDLLHSRVI 588
Query 237 GQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIR 296
GQD+AV+AVA+A+ RAGLS + P+G+FLFLG +GVGKTELAKALA E+FHSEKNLIR
Sbjct 589 GQDDAVKAVADAMVRARAGLSREGMPVGSFLFLGPTGVGKTELAKALAMEMFHSEKNLIR 648
Query 297 LDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLL 356
+DM E+ EAHS+SRLIG PPGY+G+D GGQLTEAVR++PHSVVLFDE+E H+ + +++L
Sbjct 649 IDMSEFSEAHSVSRLIGSPPGYVGHDAGGQLTEAVRRRPHSVVLFDEIEKGHQQILNIML 708
Query 357 PMLDEGHLTDTK 368
MLDEG LTD K
Sbjct 709 QMLDEGRLTDGK 720
> tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53)
Length=983
Score = 351 bits (901), Expect = 3e-96, Method: Compositional matrix adjust.
Identities = 185/363 (50%), Positives = 243/363 (66%), Gaps = 20/363 (5%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DEIHTL+GAGKA G MDA+ +LK LARG + +IGATT +EY+ ++E+D AF RRF I
Sbjct 379 DEIHTLMGAGKADGPMDAANLLKPALARGALRVIGATTRAEYRKHIERDMAFARRFVTIE 438
Query 78 VEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDEA 137
++ P +T+++L+ ++ N E HH L I D L LSD+ +K R PDKA+DL+D+A
Sbjct 439 MKEPDVAKTITMLKGIRKNLENHHKLTITDGALVAAATLSDRYIKSRQLPDKAIDLIDDA 498
Query 138 CAFRRVRHNSRRAELALLLQQQRD------------GKTHFAEEELAALQAEYAEMTEPT 185
CA ++V+ R +++ D G ++L L+AE A+ E
Sbjct 499 CAIKKVKSLRRLMNERAANEEKNDNGGSGDASSSANGDGEKKRDDLEKLEAEIAKEAEDA 558
Query 186 EERPDPEVLELQMDDVAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKRVIGQDEAVQAV 245
+ D DVA V+SL TGIPV ++TE + R+L L L +VIGQ+EAV+AV
Sbjct 559 DALTDA--------DVADVVSLRTGIPVNKLTETDRQRLLSLPASLQAQVIGQEEAVEAV 610
Query 246 ANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVEYQEA 305
ANAI RAGL+ PIG FLFLGS+GVGKTELAKAL IFH EKNLIRLDM EY E
Sbjct 611 ANAIIRSRAGLARRNAPIGTFLFLGSTGVGKTELAKALTAAIFHDEKNLIRLDMSEYSEP 670
Query 306 HSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLT 365
H+++RL+G PPGY+ +DEGGQLTEAVRQ+PHSVVLFDE+ENAH N+++ LL +LDEG LT
Sbjct 671 HTVARLVGSPPGYVSHDEGGQLTEAVRQRPHSVVLFDEIENAHPNVFAYLLQLLDEGRLT 730
Query 366 DTK 368
D +
Sbjct 731 DMR 733
> ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptidase/
ATPase; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=929
Score = 327 bits (838), Expect = 6e-89, Method: Compositional matrix adjust.
Identities = 174/377 (46%), Positives = 246/377 (65%), Gaps = 26/377 (6%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DE+HTLIGAG A G++DA+ +LK LARGE+ IGATTL EY+ ++EKD A RRFQ +
Sbjct 373 DEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEYRKHIEKDPALERRFQPVK 432
Query 78 VEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDEA 137
V P+ + T+ IL+ ++ YE HH L+ D L QLS Q + R PDKA+DL+DEA
Sbjct 433 VPEPTVDETIQILKGLRERYEIHHKLRYTDESLVAAAQLSYQYISDRFLPDKAIDLIDEA 492
Query 138 CAFRRVRHNSRRAELALLLQQQRDGKTHFAEE---------------------ELAALQA 176
+ R+RH E L ++ R E E++A+QA
Sbjct 493 GSRVRLRHAQVPEEARELEKELRQITKEKNEAVRGQDFEKAGTLRDREIELRAEVSAIQA 552
Query 177 EYAEMTEPTEERPD--PEVLELQMDDVAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKR 234
+ EM++ E + P V E D+ H++S WTGIPV +++ DE+ R+LK+ + L KR
Sbjct 553 KGKEMSKAESETGEEGPMVTE---SDIQHIVSSWTGIPVEKVSTDESDRLLKMEETLHKR 609
Query 235 VIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNL 294
+IGQDEAV+A++ AIR R GL +PI +F+F G +GVGK+ELAKALA F SE+ +
Sbjct 610 IIGQDEAVKAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAM 669
Query 295 IRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSL 354
IRLDM E+ E H++S+LIG PPGY+G EGGQLTEAVR++P++VVLFDE+E AH +++++
Sbjct 670 IRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNM 729
Query 355 LLPMLDEGHLTDTKQRA 371
+L +L++G LTD+K R
Sbjct 730 MLQILEDGRLTDSKGRT 746
> ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding /
nuclease/ nucleoside-triphosphatase/ nucleotide binding / protein
binding; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=952
Score = 317 bits (812), Expect = 5e-86, Method: Compositional matrix adjust.
Identities = 170/377 (45%), Positives = 245/377 (64%), Gaps = 26/377 (6%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DE+HTLIGAG A G++DA+ +LK LARGE+ IGATT+ EY+ ++EKD A RRFQ +
Sbjct 394 DEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTIDEYRKHIEKDPALERRFQPVK 453
Query 78 VEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDEA 137
V P+ E + IL+ ++ YE HH L+ D L QLS Q + R PDKA+DL+DEA
Sbjct 454 VPEPTVEEAIQILQGLRERYEIHHKLRYTDEALVAAAQLSHQYISDRFLPDKAIDLIDEA 513
Query 138 CAFRRVRH-----NSRRAELALL-LQQQRDGKTHFAEEELAA--------LQAEYAEM-- 181
+ R+RH +R E L + ++++ + E+A L+AE A +
Sbjct 514 GSRVRLRHAQLPEEARELEKQLRQITKEKNEAVRSQDFEMAGSHRDREIELKAEIANVLS 573
Query 182 -------TEPTEERPDPEVLELQMDDVAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKR 234
E E P V E D+ H+++ WTGIPV +++ DE++R+L++ L R
Sbjct 574 RGKEVAKAENEAEEGGPTVTE---SDIQHIVATWTGIPVEKVSSDESSRLLQMEQTLHTR 630
Query 235 VIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNL 294
VIGQDEAV+A++ AIR R GL +PI +F+F G +GVGK+ELAKALA F SE+ +
Sbjct 631 VIGQDEAVKAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAM 690
Query 295 IRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSL 354
IRLDM E+ E H++S+LIG PPGY+G EGGQLTEAVR++P+++VLFDE+E AH +++++
Sbjct 691 IRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTLVLFDEIEKAHPDVFNM 750
Query 355 LLPMLDEGHLTDTKQRA 371
+L +L++G LTD+K R
Sbjct 751 MLQILEDGRLTDSKGRT 767
> ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1);
ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=945
Score = 294 bits (753), Expect = 4e-79, Method: Compositional matrix adjust.
Identities = 160/379 (42%), Positives = 237/379 (62%), Gaps = 27/379 (7%)
Query 18 DEIHTLIGAG------KAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCR 71
DE+HTLIG+G K +G +D + +LK L RGE+ I +TTL E++ EKD A R
Sbjct 387 DEVHTLIGSGTVGRGNKGSG-LDIANLLKPSLGRGELQCIASTTLDEFRSQFEKDKALAR 445
Query 72 RFQKILVEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKAL 131
RFQ +L+ PS+E + IL ++ YEAHH+ + + V LS + + R PDKA+
Sbjct 446 RFQPVLINEPSEEDAVKILLGLREKYEAHHNCKYTMEAIDAAVYLSSRYIADRFLPDKAI 505
Query 132 DLLDEACAFRRVRHNSRRAELALLLQQQ------RDGKTHFAEEELA-----------AL 174
DL+DEA + R+ ++ E A+ + + ++ KT A E+ A+
Sbjct 506 DLIDEAGSRARIEAFRKKKEDAICILSKPPNDYWQEIKTVQAMHEVVLSSRQKQDDGDAI 565
Query 175 QAEYAEMTEPTEERP---DPEVLELQMDDVAHVLSLWTGIPVGRMTEDEATRVLKLADLL 231
E E+ E + P D E + + DD+A V S+W+GIPV ++T DE ++ L D L
Sbjct 566 SDESGELVEESSLPPAAGDDEPILVGPDDIAAVASVWSGIPVQQITADERMLLMSLEDQL 625
Query 232 SKRVIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFHSE 291
RV+GQDEAV A++ A++ R GL + +PI A LF G +GVGKTEL KALA F SE
Sbjct 626 RGRVVGQDEAVAAISRAVKRSRVGLKDPDRPIAAMLFCGPTGVGKTELTKALAANYFGSE 685
Query 292 KNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNL 351
++++RLDM EY E H++S+LIG PPGY+G +EGG LTEA+R++P +VVLFDE+E AH ++
Sbjct 686 ESMLRLDMSEYMERHTVSKLIGSPPGYVGFEEGGMLTEAIRRRPFTVVLFDEIEKAHPDI 745
Query 352 WSLLLPMLDEGHLTDTKQR 370
+++LL + ++GHLTD++ R
Sbjct 746 FNILLQLFEDGHLTDSQGR 764
> tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit
(EC:3.4.21.92); K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=900
Score = 285 bits (729), Expect = 3e-76, Method: Compositional matrix adjust.
Identities = 149/355 (41%), Positives = 218/355 (61%), Gaps = 11/355 (3%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DE H LIG G GS+DA+ +LK PL+RGEI I TT EYK Y EKD A RRF I
Sbjct 366 DEAHMLIGGGAGDGSIDAANLLKPPLSRGEIQCIAITTPKEYKKYFEKDMALSRRFHPIY 425
Query 78 VEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDEA 137
V+ PS E TL IL + +Y H ++ + ++ S Q + R PDKA+D++DE+
Sbjct 426 VDEPSDEDTLKILNGISSSYGEFHGVEYTQDSIKLALKYSKQYINDRFLPDKAIDIMDES 485
Query 138 CAFRRVRHNSRRAELALLLQQQRDGKTHFAEEELAALQAEYAEMTEPTEERPDPEVL-EL 196
+F ++++ + + +R+ + + E E E+ D VL ++
Sbjct 486 GSFAKIQYQN---------ELKREKNEIPDPSNSTSPEGENETKVEQKSEKKDQSVLGQV 536
Query 197 QMDDVAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRAGL 256
+ + VA V+S+WTGIP+ ++T E + + + L K VIGQ+EAV+ V AIR + +
Sbjct 537 KPEHVAEVMSIWTGIPLKKLTRGEMDIIRNMEEDLHKMVIGQEEAVKNVCKAIRRAKTNI 596
Query 257 SEDKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIGPPP 316
+PIG+FLF G GVGK+E+A+AL + +F E NLIR+DM EY E HSISR++G PP
Sbjct 597 KNPNRPIGSFLFCGPPGVGKSEVARALTKYLFAKE-NLIRIDMSEYTEPHSISRILGSPP 655
Query 317 GYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDTKQRA 371
GY G+D GGQLTE V+ P+SVV+FDE+E AH ++ ++LL +L++G LTD+K +
Sbjct 656 GYKGHDTGGQLTEKVKSNPYSVVMFDEIEKAHHDVLNILLQILEDGKLTDSKNQT 710
> ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding; K03695 ATP-dependent Clp protease ATP-binding
subunit ClpB
Length=968
Score = 282 bits (722), Expect = 2e-75, Method: Compositional matrix adjust.
Identities = 170/433 (39%), Positives = 247/433 (57%), Gaps = 79/433 (18%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DEIHT++GAG G+MDA +LK L RGE+ IGATTL EY+ Y+EKD A RRFQ++
Sbjct 354 DEIHTVVGAGATNGAMDAGNLLKPMLGRGELRCIGATTLDEYRKYIEKDPALERRFQQVY 413
Query 78 VEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDEA 137
V+ P+ E T+SILR ++ YE HH ++I D L + LSD+ + R PDKA+DL+DEA
Sbjct 414 VDQPTVEDTISILRGLRERYELHHGVRISDSALVEAAILSDRYISGRFLPDKAIDLVDEA 473
Query 138 CAFRRVRHNSRRAEL------ALLLQQQRDGKTHFA-----------EEELAALQAEYAE 180
A ++ S+ L + L+ +R T+ E EL L+ + AE
Sbjct 474 AAKLKMEITSKPTALDELDRSVIKLEMERLSLTNDTDKASRERLNRIETELVLLKEKQAE 533
Query 181 MTEPTE-------------ERPDPEVLELQMDDVAHVLSLWTGIPVGRMTE-----DEAT 222
+TE E E D LE+Q + + L+ + G + +EA
Sbjct 534 LTEQWEHERSVMSRLQSIKEEIDRVNLEIQQAEREYDLNRAAELKYGSLNSLQRQLNEAE 593
Query 223 RVLK------------------LADLLSK--------------------------RVIGQ 238
+ L +A+++SK RV+GQ
Sbjct 594 KELNEYLSSGKSMFREEVLGSDIAEIVSKWTGIPVSKLQQSERDKLLHLEEELHKRVVGQ 653
Query 239 DEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLD 298
+ AV AVA AI+ RAGLS+ +PI +F+F+G +GVGKTELAKALA +F++E+ L+R+D
Sbjct 654 NPAVTAVAEAIQRSRAGLSDPGRPIASFMFMGPTGVGKTELAKALASYMFNTEEALVRID 713
Query 299 MVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPM 358
M EY E H++SRLIG PPGY+G +EGGQLTE VR++P+SV+LFDE+E AH +++++ L +
Sbjct 714 MSEYMEKHAVSRLIGAPPGYVGYEEGGQLTETVRRRPYSVILFDEIEKAHGDVFNVFLQI 773
Query 359 LDEGHLTDTKQRA 371
LD+G +TD++ R
Sbjct 774 LDDGRVTDSQGRT 786
> eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity
subunit of ClpA-ClpP ATP-dependent serine protease, chaperone
activity; K03694 ATP-dependent Clp protease ATP-binding
subunit ClpA
Length=758
Score = 235 bits (600), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 137/358 (38%), Positives = 202/358 (56%), Gaps = 55/358 (15%)
Query 18 DEIHTLIGAGKAAG-SMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKI 76
DEIHT+IGAG A+G +DA+ ++K L+ G+I +IG+TT E+ EKD A RRFQKI
Sbjct 285 DEIHTIIGAGAASGGQVDAANLIKPLLSSGKIRVIGSTTYQEFSNIFEKDRALARRFQKI 344
Query 77 LVEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDE 136
+ PS E T+ I+ +K YEAHH ++ + V+L+ + + R PDKA+D++DE
Sbjct 345 DITEPSIEETVQIINGLKPKYEAHHDVRYTAKAVRAAVELAVKYINDRHLPDKAIDVIDE 404
Query 137 ACAFRRVRHNSRRAELALLLQQQRDGKTHFAEEELAALQAEYAEMTEPTEERPDPEVLEL 196
A A R+ S+R + +
Sbjct 405 AGARARLMPVSKRKK-------------------------------------------TV 421
Query 197 QMDDVAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRAGL 256
+ D+ V++ IP +++ + + L D L V GQD+A++A+ AI+ RAGL
Sbjct 422 NVADIESVVARIARIPEKSVSQSDRDTLKNLGDRLKMLVFGQDKAIEALTEAIKMARAGL 481
Query 257 SEDKKPIGAFLFLGSSGVGKTE----LAKALAEEIFHSEKNLIRLDMVEYQEAHSISRLI 312
+ KP+G+FLF G +GVGKTE L+KAL E L+R DM EY E H++SRLI
Sbjct 482 GHEHKPVGSFLFAGPTGVGKTEVTVQLSKALGIE-------LLRFDMSEYMERHTVSRLI 534
Query 313 GPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDTKQR 370
G PPGY+G D+GG LT+AV + PH+V+L DE+E AH +++++LL ++D G LTD R
Sbjct 535 GAPPGYVGFDQGGLLTDAVIKHPHAVLLLDEIEKAHPDVFNILLQVMDNGTLTDNNGR 592
> ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN
101); ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=911
Score = 214 bits (546), Expect = 4e-55, Method: Compositional matrix adjust.
Identities = 110/212 (51%), Positives = 146/212 (68%), Gaps = 7/212 (3%)
Query 160 RDGKTHFAEEELAALQAEYAEMTEPTEERPDPEVLELQMDDVAHVLSLWTGIPVGRMTED 219
R G E +A L+ +E E PE +A V+S WTGIPV R+ ++
Sbjct 503 RYGAIQEVESAIAQLEGTSSEENVMLTENVGPE-------HIAEVVSRWTGIPVTRLGQN 555
Query 220 EATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTEL 279
E R++ LAD L KRV+GQ++AV AV+ AI RAGL ++P G+FLFLG +GVGKTEL
Sbjct 556 EKERLIGLADRLHKRVVGQNQAVNAVSEAILRSRAGLGRPQQPTGSFLFLGPTGVGKTEL 615
Query 280 AKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVV 339
AKALAE++F E L+R+DM EY E HS+SRLIG PPGY+G++EGGQLTEAVR++P+ V+
Sbjct 616 AKALAEQLFDDENLLVRIDMSEYMEQHSVSRLIGAPPGYVGHEEGGQLTEAVRRRPYCVI 675
Query 340 LFDEVENAHKNLWSLLLPMLDEGHLTDTKQRA 371
LFDEVE AH +++ LL +LD+G LTD + R
Sbjct 676 LFDEVEKAHVAVFNTLLQVLDDGRLTDGQGRT 707
Score = 141 bits (355), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 72/135 (53%), Positives = 94/135 (69%), Gaps = 0/135 (0%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DEIH ++GAGK GSMDA+ + K LARG++ IGATTL EY+ Y+EKDAAF RRFQ++
Sbjct 279 DEIHLVLGAGKTEGSMDAANLFKPMLARGQLRCIGATTLEEYRKYVEKDAAFERRFQQVY 338
Query 78 VEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDEA 137
V PS T+SILR +K YE HH ++I D L QLS + + R PDKA+DL+DEA
Sbjct 339 VAEPSVPDTISILRGLKEKYEGHHGVRIQDRALINAAQLSARYITGRHLPDKAIDLVDEA 398
Query 138 CAFRRVRHNSRRAEL 152
CA RV+ +S+ E+
Sbjct 399 CANVRVQLDSQPEEI 413
> eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation
chaperone; K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=857
Score = 213 bits (541), Expect = 2e-54, Method: Compositional matrix adjust.
Identities = 98/172 (56%), Positives = 135/172 (78%), Gaps = 0/172 (0%)
Query 200 DVAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRAGLSED 259
++A VL+ WTGIPV RM E E ++L++ L RVIGQ+EAV AV+NAIR RAGL++
Sbjct 535 EIAEVLARWTGIPVSRMMESEREKLLRMEQELHHRVIGQNEAVDAVSNAIRRSRAGLADP 594
Query 260 KKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYM 319
+PIG+FLFLG +GVGKTEL KALA +F S++ ++R+DM E+ E HS+SRL+G PPGY+
Sbjct 595 NRPIGSFLFLGPTGVGKTELCKALANFMFDSDEAMVRIDMSEFMEKHSVSRLVGAPPGYV 654
Query 320 GNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDTKQRA 371
G +EGG LTEAVR++P+SV+L DEVE AH +++++LL +LD+G LTD + R
Sbjct 655 GYEEGGYLTEAVRRRPYSVILLDEVEKAHPDVFNILLQVLDDGRLTDGQGRT 706
Score = 140 bits (354), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 71/135 (52%), Positives = 94/135 (69%), Gaps = 0/135 (0%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DE+HT++GAGKA G+MDA MLK LARGE+ +GATTL EY+ Y+EKDAA RRFQK+
Sbjct 278 DELHTMVGAGKADGAMDAGNMLKPALARGELHCVGATTLDEYRQYIEKDAALERRFQKVF 337
Query 78 VEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDEA 137
V PS E T++ILR +K YE HH +QI D + LS + + R PDKA+DL+DEA
Sbjct 338 VAEPSVEDTIAILRGLKERYELHHHVQITDPAIVAAATLSHRYIADRQLPDKAIDLIDEA 397
Query 138 CAFRRVRHNSRRAEL 152
+ R++ +S+ EL
Sbjct 398 ASSIRMQIDSKPEEL 412
> ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding
Length=964
Score = 212 bits (540), Expect = 2e-54, Method: Compositional matrix adjust.
Identities = 93/172 (54%), Positives = 137/172 (79%), Gaps = 0/172 (0%)
Query 200 DVAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRAGLSED 259
D+A ++S WTGIP+ + + E +++ L ++L RVIGQD AV++VA+AIR RAGLS+
Sbjct 620 DIAEIVSKWTGIPLSNLQQSEREKLVMLEEVLHHRVIGQDMAVKSVADAIRRSRAGLSDP 679
Query 260 KKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYM 319
+PI +F+F+G +GVGKTELAKALA +F++E ++R+DM EY E HS+SRL+G PPGY+
Sbjct 680 NRPIASFMFMGPTGVGKTELAKALAGYLFNTENAIVRVDMSEYMEKHSVSRLVGAPPGYV 739
Query 320 GNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDTKQRA 371
G +EGGQLTE VR++P+SVVLFDE+E AH +++++LL +LD+G +TD++ R
Sbjct 740 GYEEGGQLTEVVRRRPYSVVLFDEIEKAHPDVFNILLQLLDDGRITDSQGRT 791
Score = 132 bits (331), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 68/135 (50%), Positives = 90/135 (66%), Gaps = 0/135 (0%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DEIHT++GAG G+MDAS +LK L RGE+ IGATTL+EY+ Y+EKD A RRFQ++L
Sbjct 359 DEIHTVVGAGAMDGAMDASNLLKPMLGRGELRCIGATTLTEYRKYIEKDPALERRFQQVL 418
Query 78 VEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDEA 137
PS E T+SILR ++ YE HH + I D L L+D+ + R PDKA+DL+DEA
Sbjct 419 CVQPSVEDTISILRGLRERYELHHGVTISDSALVSAAVLADRYITERFLPDKAIDLVDEA 478
Query 138 CAFRRVRHNSRRAEL 152
A ++ S+ EL
Sbjct 479 GAKLKMEITSKPTEL 493
> tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=985
Score = 212 bits (540), Expect = 2e-54, Method: Compositional matrix adjust.
Identities = 90/169 (53%), Positives = 137/169 (81%), Gaps = 0/169 (0%)
Query 199 DDVAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRAGLSE 258
+D+A+V+S WTGIP+ ++ + + ++L+L D L KR+IGQ EA+ AV NA++ R G+++
Sbjct 660 EDIANVVSKWTGIPLNKLIKSQKEKILQLNDELHKRIIGQQEAIDAVVNAVQRSRVGMND 719
Query 259 DKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIGPPPGY 318
KKPI A +FLG +GVGKTEL+KA+AE++F SE+ +IR DM EY E HS+S+L+G PPGY
Sbjct 720 PKKPIAALMFLGPTGVGKTELSKAIAEQLFDSEEAIIRFDMSEYMEKHSVSKLVGAPPGY 779
Query 319 MGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDT 367
+G ++GG LTEA+R+KP+S++LFDE+E AH +++++LL +LD+G LTD+
Sbjct 780 IGYEQGGLLTEAIRRKPYSILLFDEIEKAHSDVYNILLQVLDDGRLTDS 828
Score = 135 bits (339), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 69/142 (48%), Positives = 99/142 (69%), Gaps = 0/142 (0%)
Query 14 VVGRDEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRF 73
V+ DEIHTL+GAG++ GS+DA +LK LARGE+ IGATTL EY+ +EKD A RRF
Sbjct 376 VMFIDEIHTLVGAGESQGSLDAGNILKPMLARGELRCIGATTLQEYRQKIEKDKALERRF 435
Query 74 QKILVEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDL 133
Q I ++ P+ E T++ILR +K YE HH ++I D L Q V LS++ + R PDKA+DL
Sbjct 436 QPIYIDEPNIEETINILRGLKERYEVHHGVRILDSTLIQAVLLSNRYITDRYLPDKAIDL 495
Query 134 LDEACAFRRVRHNSRRAELALL 155
+DEA A +++ +S+ +L ++
Sbjct 496 IDEAAAKLKIQLSSKPLQLDII 517
> bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=931
Score = 212 bits (539), Expect = 3e-54, Method: Compositional matrix adjust.
Identities = 91/176 (51%), Positives = 134/176 (76%), Gaps = 0/176 (0%)
Query 195 ELQMDDVAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRA 254
E+ DD+A V+S WTGIPV ++ + ++L + D L KR+IGQDEAV V A++ R
Sbjct 601 EVSRDDIASVVSRWTGIPVNKLIRSQRDKILHIGDELRKRIIGQDEAVDIVTRAVQRSRV 660
Query 255 GLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIGP 314
G+++ K+PI +FLG +GVGKTEL KA+AE++F +++ +IR DM EY E HS+SRL+G
Sbjct 661 GMNDPKRPIAGLMFLGPTGVGKTELCKAIAEQLFDTDEAIIRFDMSEYMEKHSVSRLVGA 720
Query 315 PPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDTKQR 370
PPGY+G D+GG LTEAVR++P+S+VLFDE+E AH ++++++L +LD+G LTD+ R
Sbjct 721 PPGYIGYDQGGLLTEAVRRRPYSIVLFDEIEKAHPDVFNIMLQLLDDGRLTDSSGR 776
Score = 135 bits (340), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 69/135 (51%), Positives = 94/135 (69%), Gaps = 0/135 (0%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DEIHT++GAG A G+MDA +LK LARGE+ IGATTL EY+ +EKD A RRFQ +
Sbjct 320 DEIHTVVGAGDAQGAMDAGNILKPMLARGELRCIGATTLQEYRQRIEKDKALERRFQPVY 379
Query 78 VEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDEA 137
V+ PS E T+SILR ++ YE HH ++I D L + QLSD+ + R PDKA+DL+DEA
Sbjct 380 VDQPSVEETISILRGLRERYEVHHGVRILDSALVEAAQLSDRYITDRFLPDKAIDLVDEA 439
Query 138 CAFRRVRHNSRRAEL 152
A +++ +S+ +L
Sbjct 440 AARLKIQLSSKPIQL 454
> tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53)
Length=921
Score = 207 bits (528), Expect = 4e-53, Method: Compositional matrix adjust.
Identities = 97/171 (56%), Positives = 128/171 (74%), Gaps = 0/171 (0%)
Query 201 VAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRAGLSEDK 260
+A V+ WT IPV ++T+ E R L L L+++VIGQ +AV+AV AI AGLS
Sbjct 538 IADVVHRWTNIPVQKLTQTETERFLTLGKSLAEQVIGQPQAVEAVTQAILRSAAGLSRRN 597
Query 261 KPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYMG 320
+PIG+FLFLG +GVGKTEL K +AE +F S++ L+R DM EY E HS+SRLIG PPGY+G
Sbjct 598 RPIGSFLFLGPTGVGKTELCKRVAESLFDSKERLVRFDMSEYMEQHSVSRLIGAPPGYVG 657
Query 321 NDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDTKQRA 371
+DEGGQLTE +R+ P+SVVLFDEVE AH +W++LL +LD+G LTD++ R
Sbjct 658 HDEGGQLTEEIRRNPYSVVLFDEVEKAHSQVWNVLLQVLDDGRLTDSQGRT 708
Score = 146 bits (368), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 96/233 (41%), Positives = 137/233 (58%), Gaps = 19/233 (8%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DEIH ++GAGK G++DA+ +LK LARGE+ IGATTL EY+ Y+EKDAAF RRFQ++
Sbjct 276 DEIHVILGAGKTEGALDAANLLKPMLARGELRCIGATTLDEYRKYVEKDAAFERRFQQVH 335
Query 78 VEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDEA 137
V PS + T+SILR +K Y +HH ++I D L + QL+D+ + R PDKA+DL+DEA
Sbjct 336 VREPSVQATISILRGLKDRYASHHGVRILDSALVEAAQLADRYITSRFLPDKAIDLMDEA 395
Query 138 CAFRRVRHNSRRAELALLLQQQRDGKTHFAEEELAALQAEYAEMTEPTEERPDPEVLELQ 197
CA RV+ +S+ + +L +Q+ E EL AL+ E +P ++ EV E
Sbjct 396 CAIARVQVDSKPEAVDVLERQKVQ-----LEVELLALEKE----KDPASQKRLAEVKE-H 445
Query 198 MDDVAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKRVIGQDEAVQAVANAIR 250
+ +VA L P+ + E R+ +L L K QDE + A R
Sbjct 446 LGEVADALR-----PLYLQYQQEKARIDELGKLAQK----QDELKAKIERAQR 489
> tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.21.53);
K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=898
Score = 207 bits (526), Expect = 8e-53, Method: Compositional matrix adjust.
Identities = 92/174 (52%), Positives = 135/174 (77%), Gaps = 0/174 (0%)
Query 195 ELQMDDVAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRA 254
E+ +DD+A V+++WTGIPV R+ + E ++L L L +RV+GQD AVQ VA AI+ RA
Sbjct 688 EVTVDDIATVVAMWTGIPVTRLKQSEKEKLLNLEKDLHRRVVGQDHAVQVVAEAIQRSRA 747
Query 255 GLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIGP 314
GL++ +PI + FLG +GVGKTEL ++LAE +F SE ++++DM EY E H+ISRL+G
Sbjct 748 GLNDPNRPIASLFFLGPTGVGKTELCRSLAELMFDSEDAMVKIDMSEYMEKHTISRLLGA 807
Query 315 PPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDTK 368
PPGY+G ++GGQLT+ VR+KP+SV+LFDE+E AH +++++LL +LD+G +TD K
Sbjct 808 PPGYVGYEQGGQLTDEVRKKPYSVILFDEMEKAHPDVFNVLLQILDDGRVTDGK 861
Score = 111 bits (278), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 64/137 (46%), Positives = 86/137 (62%), Gaps = 2/137 (1%)
Query 18 DEIHTLIGAGKAAGSMDAS--QMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQK 75
DEIHT++GAG MLK LARGE IGATT +EY+ Y+EKD A RRFQK
Sbjct 425 DEIHTVVGAGAGGEGGAMDAGNMLKPMLARGEFRCIGATTTNEYRQYIEKDKALERRFQK 484
Query 76 ILVEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLD 135
+LVE P T+SILR +K YE HH ++I D L + L+ + + R PDKA+DL+D
Sbjct 485 VLVEEPQVSETISILRGLKDRYEVHHGVRILDSALVEAANLAHRYISDRFLPDKAIDLVD 544
Query 136 EACAFRRVRHNSRRAEL 152
EA A +++ +S+ +L
Sbjct 545 EAAARLKIQVSSKPIQL 561
> sce:YDR258C HSP78; Hsp78p
Length=811
Score = 201 bits (511), Expect = 5e-51, Method: Compositional matrix adjust.
Identities = 92/169 (54%), Positives = 130/169 (76%), Gaps = 0/169 (0%)
Query 199 DDVAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRAGLSE 258
DD++ V++ TGIP + + + R+L + + L +RV+GQDEA+ A+++A+R RAGL+
Sbjct 470 DDISKVVAKMTGIPTETVMKGDKDRLLYMENSLKERVVGQDEAIAAISDAVRLQRAGLTS 529
Query 259 DKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIGPPPGY 318
+K+PI +F+FLG +G GKTEL KALAE +F E N+IR DM E+QE H++SRLIG PPGY
Sbjct 530 EKRPIASFMFLGPTGTGKTELTKALAEFLFDDESNVIRFDMSEFQEKHTVSRLIGAPPGY 589
Query 319 MGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDT 367
+ ++ GGQLTEAVR+KP++VVLFDE E AH ++ LLL +LDEG LTD+
Sbjct 590 VLSESGGQLTEAVRRKPYAVVLFDEFEKAHPDVSKLLLQVLDEGKLTDS 638
Score = 134 bits (338), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 81/195 (41%), Positives = 117/195 (60%), Gaps = 8/195 (4%)
Query 14 VVGRDEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRF 73
+V DE+H L+G GK GSMDAS +LK LARG + I ATTL E+K+ +EKD A RRF
Sbjct 211 IVFIDEVHMLLGLGKTDGSMDASNILKPKLARG-LRCISATTLDEFKI-IEKDPALSRRF 268
Query 74 QKILVEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDL 133
Q IL+ PS T+SILR +K YE HH ++I D L LS++ + R PDKA+DL
Sbjct 269 QPILLNEPSVSDTISILRGLKERYEVHHGVRITDTALVSAAVLSNRYITDRFLPDKAIDL 328
Query 134 LDEACAFRRVRHNSRRAELALLLQQQRDGKTHFAEEELAALQAEYAEMTEPTEERPDPEV 193
+DEACA R++H S+ E+ Q+ D + EL +L+ E ++ E + +
Sbjct 329 VDEACAVLRLQHESKPDEI-----QKLDRAIMKIQIELESLKKETDPVSVERREALEKD- 382
Query 194 LELQMDDVAHVLSLW 208
LE++ D++ + +W
Sbjct 383 LEMKNDELNRLTKIW 397
> tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.21.53)
Length=929
Score = 200 bits (509), Expect = 8e-51, Method: Compositional matrix adjust.
Identities = 92/169 (54%), Positives = 129/169 (76%), Gaps = 0/169 (0%)
Query 199 DDVAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRAGLSE 258
+D+A V+ WTGIPV R+ E E ++L L L+ RVIGQ+E V++VA AI+ RAGL +
Sbjct 560 EDIAQVVGSWTGIPVSRLVEGEREKLLGLEKALNDRVIGQEEGVRSVAEAIQRSRAGLCD 619
Query 259 DKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIGPPPGY 318
+PI + +FLG +GVGKTEL KALA ++F +E+ L+R DM EY E HS +RLIG PPGY
Sbjct 620 PNRPIASLVFLGPTGVGKTELCKALARQLFDTEEALLRFDMSEYMEKHSTARLIGAPPGY 679
Query 319 MGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDT 367
+G D+GGQLTEAVR++P+SV+LFDE+E AH ++++ L +L++G LTD+
Sbjct 680 IGFDKGGQLTEAVRRRPYSVLLFDEMEKAHPEVFNIFLQILEDGILTDS 728
Score = 122 bits (307), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 67/136 (49%), Positives = 87/136 (63%), Gaps = 1/136 (0%)
Query 18 DEIHTLIGAGKAAGS-MDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKI 76
DEIH ++GAG A S MDA +LK LARGE+ IGATTL EY+ Y+EKD A RRFQ +
Sbjct 295 DEIHMVVGAGSAGESGMDAGNILKPMLARGELRCIGATTLDEYRKYIEKDKALERRFQVV 354
Query 77 LVEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDE 136
LV+ P E LSILR +K YE HH + I D L LS++ ++ R PDKA+DL+DE
Sbjct 355 LVDEPRVEDALSILRGLKERYEMHHGVSIRDSALVAACVLSNRYIQDRFLPDKAIDLIDE 414
Query 137 ACAFRRVRHNSRRAEL 152
A + ++ S+ L
Sbjct 415 AASKIKIEVTSKPTRL 430
> bbo:BBOV_III008980 17.m07783; Clp amino terminal domain containing
protein; K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=1005
Score = 190 bits (483), Expect = 8e-48, Method: Compositional matrix adjust.
Identities = 93/213 (43%), Positives = 140/213 (65%), Gaps = 3/213 (1%)
Query 161 DGKTHFAEEELAALQAEYAEMTEPTEERPDPE--VLELQMDDVAHVLSLWTGIPVGRMTE 218
DG +F ++ + + E T D + E+ + VA V+S WTG+P+ ++T+
Sbjct 585 DGSINFPATDMISEENYDTEYTSNASWDSDSTETLCEVNTEHVAEVVSNWTGVPLKKLTQ 644
Query 219 DEATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTE 278
E + L L K V+G +EAV+ +A AIR + + ++PIG+FLF G GVGK+E
Sbjct 645 GEIEAIRNLEQELHKSVVGHEEAVKNIAKAIRRAKTNIKNPERPIGSFLFCGPPGVGKSE 704
Query 279 LAKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSV 338
+AK L + +F +E NLI+LDM EY E HSISR++G PPGY G+D GGQLTE +R+ P+SV
Sbjct 705 IAKTLTKLMF-TEDNLIKLDMSEYNEPHSISRILGSPPGYKGHDSGGQLTEKLRKNPYSV 763
Query 339 VLFDEVENAHKNLWSLLLPMLDEGHLTDTKQRA 371
V+FDE+E AH+N+ ++LL +L++G LTD+K +
Sbjct 764 VMFDEIEKAHRNVLNILLQILEDGKLTDSKNQT 796
Score = 102 bits (254), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 54/127 (42%), Positives = 76/127 (59%), Gaps = 0/127 (0%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DE H L+GAG G++DA+ +LK LARGEI I TT EY+ + EKDAA CRRFQ I
Sbjct 381 DEAHMLVGAGAGEGALDAANLLKPTLARGEIQCIAITTPKEYQKHFEKDAALCRRFQPIH 440
Query 78 VEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDEA 137
V+ PS + T IL H+++ ++ ++ S Q + R PDKA+D+LDEA
Sbjct 441 VKEPSDKDTQIILNATAEACGRFHNVKYNMDAVAAALKYSKQFIPERYLPDKAIDILDEA 500
Query 138 CAFRRVR 144
+ ++R
Sbjct 501 GSLAKIR 507
> pfa:PF08_0063 ClpB protein, putative
Length=1070
Score = 187 bits (474), Expect = 9e-47, Method: Compositional matrix adjust.
Identities = 82/174 (47%), Positives = 131/174 (75%), Gaps = 0/174 (0%)
Query 195 ELQMDDVAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRA 254
E+ +D+ +++S+ TGI + ++ + E ++L L + L K++IGQD+AV+ V A++ R
Sbjct 741 EVTSEDIVNIVSMSTGIRLNKLLKSEKEKILNLENELHKQIIGQDDAVKVVTKAVQRSRV 800
Query 255 GLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIGP 314
G++ K+PI + +FLG +GVGKTEL+K LA+ +F + + +I DM EY E HSIS+LIG
Sbjct 801 GMNNPKRPIASLMFLGPTGVGKTELSKVLADVLFDTPEAVIHFDMSEYMEKHSISKLIGA 860
Query 315 PPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDTK 368
PGY+G ++GG LT+AVR+KP+S++LFDE+E AH ++++LLL ++DEG L+DTK
Sbjct 861 APGYVGYEQGGLLTDAVRKKPYSIILFDEIEKAHPDVYNLLLRVIDEGKLSDTK 914
Score = 135 bits (341), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 72/146 (49%), Positives = 102/146 (69%), Gaps = 1/146 (0%)
Query 14 VVGRDEIHTLIGAGKAA-GSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRR 72
V+ DEIHT++GAG A G++DA +LK LARGE+ IGATT+SEY+ ++EKD A RR
Sbjct 427 VMFIDEIHTVVGAGAVAEGALDAGNILKPMLARGELRCIGATTVSEYRQFIEKDKALERR 486
Query 73 FQKILVEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALD 132
FQ+ILVE PS + T+SILR +K YE HH ++I D L Q LSD+ + R PDKA+D
Sbjct 487 FQQILVEQPSVDETISILRGLKERYEVHHGVRILDSALVQAAVLSDRYISYRFLPDKAID 546
Query 133 LLDEACAFRRVRHNSRRAELALLLQQ 158
L+DEA + +++ +S+ +L + +Q
Sbjct 547 LIDEAASNLKIQLSSKPIQLENIEKQ 572
> pfa:PF11_0175 heat shock protein 101, putative
Length=906
Score = 181 bits (458), Expect = 6e-45, Method: Compositional matrix adjust.
Identities = 81/170 (47%), Positives = 128/170 (75%), Gaps = 0/170 (0%)
Query 201 VAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRAGLSEDK 260
V+++ +G+P+G ++ + + LKL + LSK +IG ++ ++++++A+ G+ + +
Sbjct 569 VSYIYLRDSGMPLGSLSFESSKGALKLYNSLSKSIIGNEDIIKSLSDAVVKAATGMKDPE 628
Query 261 KPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYMG 320
KPIG FLFLG +GVGKTELAK LA E+F+S+ NLIR++M E+ EAHS+S++ G PPGY+G
Sbjct 629 KPIGTFLFLGPTGVGKTELAKTLAIELFNSKDNLIRVNMSEFTEAHSVSKITGSPPGYVG 688
Query 321 NDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDTKQR 370
+ GQLTEAVR+KPHSVVLFDE+E AH +++ +LL +L +G++ D +R
Sbjct 689 FSDSGQLTEAVREKPHSVVLFDELEKAHADVFKVLLQILGDGYINDNHRR 738
Score = 136 bits (342), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 65/127 (51%), Positives = 92/127 (72%), Gaps = 0/127 (0%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DEIH L+GAGKA G DA+ +LK L++GEI LIGATT++EY+ ++E +AF RRF+KIL
Sbjct 307 DEIHLLLGAGKAEGGTDAANLLKPVLSKGEIKLIGATTIAEYRKFIESCSAFERRFEKIL 366
Query 78 VEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDEA 137
VE PS + T+ ILR +K YE + + I D L ++SD+ +K R PDKA+DLL++A
Sbjct 367 VEPPSVDMTVKILRSLKSKYENFYGINITDKALVAAAKISDRFIKDRYLPDKAIDLLNKA 426
Query 138 CAFRRVR 144
C+F +V+
Sbjct 427 CSFLQVQ 433
> sce:YLL026W HSP104; Heat shock protein that cooperates with
Ydj1p (Hsp40) and Ssa1p (Hsp70) to refold and reactivate previously
denatured, aggregated proteins; responsive to stresses
including: heat, ethanol, and sodium arsenite; involved in
[PSI+] propagation
Length=908
Score = 155 bits (391), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 75/167 (44%), Positives = 118/167 (70%), Gaps = 1/167 (0%)
Query 199 DDVAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRAGLSE 258
D ++ + TGIPV +++E E +++ + LS V+GQ +A++AV+NA+R R+GL+
Sbjct 544 DTISETAARLTGIPVKKLSESENEKLIHMERDLSSEVVGQMDAIKAVSNAVRLSRSGLAN 603
Query 259 DKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIGPPPGY 318
++P +FLFLG SG GKTELAK +A +F+ E +IR+D E E +++S+L+G GY
Sbjct 604 PRQP-ASFLFLGLSGSGKTELAKKVAGFLFNDEDMMIRVDCSELSEKYAVSKLLGTTAGY 662
Query 319 MGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDEGHLT 365
+G DEGG LT ++ KP+SV+LFDEVE AH ++ +++L MLD+G +T
Sbjct 663 VGYDEGGFLTNQLQYKPYSVLLFDEVEKAHPDVLTVMLQMLDDGRIT 709
Score = 113 bits (282), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 64/135 (47%), Positives = 87/135 (64%), Gaps = 4/135 (2%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DEIH L+G GK DA+ +LK L+RG++ +IGATT +EY+ +EKD AF RRFQKI
Sbjct 284 DEIHMLMGNGKD----DAANILKPALSRGQLKVIGATTNNEYRSIVEKDGAFERRFQKIE 339
Query 78 VEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDEA 137
V PS +T++ILR ++ YE HH ++I D L QL+ + L R PD ALDL+D +
Sbjct 340 VAEPSVRQTVAILRGLQPKYEIHHGVRILDSALVTAAQLAKRYLPYRRLPDSALDLVDIS 399
Query 138 CAFRRVRHNSRRAEL 152
CA V +S+ EL
Sbjct 400 CAGVAVARDSKPEEL 414
> dre:100331587 suppressor of K+ transport defect 3-like
Length=409
Score = 151 bits (381), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 76/155 (49%), Positives = 106/155 (68%), Gaps = 2/155 (1%)
Query 215 RMTEDEATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGV 274
R E E R L L + +IGQ+ A+ VA+AIR G +++ P+ FLFLGSSG+
Sbjct 38 RKREAEERRKFPLERRLKEHIIGQEGAINTVASAIRRKENGWYDEEHPL-VFLFLGSSGI 96
Query 275 GKTELAKALAEEIFHS-EKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQ 333
GKTELAK +A + +K IR+DM E+QE H +++ IG PPGY+G+DEGGQLT+ ++Q
Sbjct 97 GKTELAKQVARYMHKDIKKGFIRMDMSEFQEKHEVAKFIGSPPGYVGHDEGGQLTKQLKQ 156
Query 334 KPHSVVLFDEVENAHKNLWSLLLPMLDEGHLTDTK 368
P +VVLFDEVE AH ++ +++L + DEG LTD K
Sbjct 157 SPSAVVLFDEVEKAHPDVLTVMLQLFDEGRLTDGK 191
> ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosphatase/
nucleotide binding / protein binding
Length=623
Score = 149 bits (376), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 70/121 (57%), Positives = 94/121 (77%), Gaps = 0/121 (0%)
Query 199 DDVAHVLSLWTGIPVGRMTEDEATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRAGLSE 258
+++A V+S WTGIPV R+ ++E R++ LAD L +RV+GQDEAV+AVA AI R GL
Sbjct 500 ENIAEVVSRWTGIPVTRLDQNEKKRLISLADKLHERVVGQDEAVKAVAAAILRSRVGLGR 559
Query 259 DKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIGPPPGY 318
++P G+FLFLG +GVGKTELAKALAE++F SE L+RLDM EY + S+++LIG PPGY
Sbjct 560 PQQPSGSFLFLGPTGVGKTELAKALAEQLFDSENLLVRLDMSEYNDKFSVNKLIGAPPGY 619
Query 319 M 319
+
Sbjct 620 V 620
Score = 133 bits (334), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 66/122 (54%), Positives = 88/122 (72%), Gaps = 0/122 (0%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DEIH +GA KA+GS DA+++LK LARG++ IGATTL EY+ ++EKDAAF RRFQ++
Sbjct 244 DEIHMALGACKASGSTDAAKLLKPMLARGQLRFIGATTLEEYRTHVEKDAAFERRFQQVF 303
Query 78 VEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDEA 137
V PS T+SILR +K YE HH ++I D L QLS++ + R PDKA+DL+DE+
Sbjct 304 VAEPSVPDTISILRGLKEKYEGHHGVRIQDRALVLSAQLSERYITGRRLPDKAIDLVDES 363
Query 138 CA 139
CA
Sbjct 364 CA 365
> mmu:20480 Clpb, AL118244, Skd3; ClpB caseinolytic peptidase
B homolog (E. coli); K03695 ATP-dependent Clp protease ATP-binding
subunit ClpB
Length=677
Score = 143 bits (361), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 69/139 (49%), Positives = 101/139 (72%), Gaps = 2/139 (1%)
Query 231 LSKRVIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFH- 289
L + +IGQ+ A+ V AIR G +++ P+ FLFLGSSG+GKTELAK A+ +
Sbjct 313 LKEHIIGQESAIATVGAAIRRKENGWYDEEHPL-VFLFLGSSGIGKTELAKQTAKYMHKD 371
Query 290 SEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHK 349
++K IRLDM E+QE H +++ IG PPGY+G++EGGQLT+ ++Q P++VVLFDEV+ AH
Sbjct 372 AKKGFIRLDMSEFQERHEVAKFIGSPPGYIGHEEGGQLTKKLKQCPNAVVLFDEVDKAHP 431
Query 350 NLWSLLLPMLDEGHLTDTK 368
++ +++L + DEG LTD K
Sbjct 432 DVLTIMLQLFDEGRLTDGK 450
> hsa:81570 CLPB, FLJ13152, HSP78, SKD3; ClpB caseinolytic peptidase
B homolog (E. coli); K03695 ATP-dependent Clp protease
ATP-binding subunit ClpB
Length=707
Score = 143 bits (361), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 69/139 (49%), Positives = 101/139 (72%), Gaps = 2/139 (1%)
Query 231 LSKRVIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFH- 289
L + +IGQ+ A+ V AIR G +++ P+ FLFLGSSG+GKTELAK A+ +
Sbjct 343 LKEHIIGQESAIATVGAAIRRKENGWYDEEHPL-VFLFLGSSGIGKTELAKQTAKYMHKD 401
Query 290 SEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHK 349
++K IRLDM E+QE H +++ IG PPGY+G++EGGQLT+ ++Q P++VVLFDEV+ AH
Sbjct 402 AKKGFIRLDMSEFQERHEVAKFIGSPPGYVGHEEGGQLTKKLKQCPNAVVLFDEVDKAHP 461
Query 350 NLWSLLLPMLDEGHLTDTK 368
++ +++L + DEG LTD K
Sbjct 462 DVLTIMLQLFDEGRLTDGK 480
> bbo:BBOV_V000160 clpC
Length=551
Score = 140 bits (354), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 62/149 (41%), Positives = 95/149 (63%), Gaps = 2/149 (1%)
Query 220 EATRVLKLADLLSKRVIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTEL 279
+ V +L++K + GQD + ++ + N + KPIG+FL G SG GKTE+
Sbjct 262 NTSLVYNFYNLINKFIFGQDLITKKISTYLLNF--NFNNKIKPIGSFLLCGPSGTGKTEI 319
Query 280 AKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVV 339
K + ++ S+ NL++ DM EY+E+HS+S+LIG PPGY+G++ GG L + +V
Sbjct 320 VKLITNYLYKSQTNLLQFDMSEYKESHSVSKLIGAPPGYVGHESGGNLINKINSVESPIV 379
Query 340 LFDEVENAHKNLWSLLLPMLDEGHLTDTK 368
LFDE+E A KN++S+ L +LDEG LTD+K
Sbjct 380 LFDEIEKADKNIFSIFLQILDEGLLTDSK 408
> pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP-dependent
Clp protease ATP-binding subunit ClpB
Length=1341
Score = 127 bits (319), Expect = 8e-29, Method: Composition-based stats.
Identities = 60/134 (44%), Positives = 92/134 (68%), Gaps = 1/134 (0%)
Query 235 VIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNL 294
+IGQ++ + ++ + + + KPIG L GSSGVGKT A+ +++ +F +E NL
Sbjct 933 IIGQEKVIDILSKYLFKAITNIKDPNKPIGTLLLCGSSGVGKTLCAQVISKYLF-NEDNL 991
Query 295 IRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSL 354
I ++M EY + HS+S+L G PGY+G EGG+LTE+V++KP S++LFDE+E AH + +
Sbjct 992 IVINMSEYIDKHSVSKLFGSYPGYVGYKEGGELTESVKKKPFSIILFDEIEKAHSEVLHV 1051
Query 355 LLPMLDEGHLTDTK 368
LL +LD G LTD+K
Sbjct 1052 LLQILDNGLLTDSK 1065
Score = 110 bits (274), Expect = 1e-23, Method: Composition-based stats.
Identities = 55/153 (35%), Positives = 89/153 (58%), Gaps = 1/153 (0%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DEIH ++GAG GS+DAS +LK L+ + IG TT EY ++E D A RRF +
Sbjct 622 DEIHVIVGAGSGEGSLDASNLLKPFLSSDNLQCIGTTTFQEYSKFIENDKALRRRFNCVT 681
Query 78 VEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDEA 137
+ + + T +L+K+K NYE +H++ D L IV L++ L FPDKA+D+LDEA
Sbjct 682 INPFTSKETYKLLKKIKYNYEKYHNIYYTDDSLKSIVSLTEDYLPTANFPDKAIDILDEA 741
Query 138 CAFRRVRHNSRRAELALLLQQQRDGKTHFAEEE 170
++++++ + + L ++ R + H +E
Sbjct 742 GVYQKIKY-EKFMKQKLRAERLRKIRIHMNTQE 773
> bbo:BBOV_V000150 clpC
Length=541
Score = 125 bits (315), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 58/124 (46%), Positives = 81/124 (65%), Gaps = 1/124 (0%)
Query 242 VQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVE 301
++ V +I N + KP+ +FLF G SG GKTELAK +F S K LI+L+M E
Sbjct 258 IEPVVKSI-NKSFYIPSKTKPLSSFLFCGPSGAGKTELAKIFTYSLFKSTKQLIKLNMSE 316
Query 302 YQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPMLDE 361
Y EAHSIS+++G PPGY+G +E V+ P+ V+LFDE+E AHK++ L+L +L+E
Sbjct 317 YMEAHSISKILGSPPGYVGYNENNDFINKVKSMPNCVILFDEIEKAHKSINDLMLQLLEE 376
Query 362 GHLT 365
G LT
Sbjct 377 GKLT 380
> tpv:TP05_0024 clpC; molecular chaperone
Length=529
Score = 123 bits (309), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 60/137 (43%), Positives = 87/137 (63%), Gaps = 0/137 (0%)
Query 229 DLLSKRVIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIF 288
+ L ++ QDEAV++V I+ ++ KP+G++L G SG GKTELAK L IF
Sbjct 237 NYLKSKLFDQDEAVESVLYRIKKLSNANIDETKPLGSWLLCGPSGTGKTELAKILCYTIF 296
Query 289 HSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAH 348
+S NLI+++M EY E H++S+LIG PPGY G E L+ + V+LFDE+E AH
Sbjct 297 NSHDNLIKINMAEYVEKHAVSKLIGSPPGYSGYGEDTILSTKFKTGSSFVILFDEIEKAH 356
Query 349 KNLWSLLLPMLDEGHLT 365
++ L+L +LD+G LT
Sbjct 357 TSITDLMLQILDKGKLT 373
> bbo:BBOV_I001700 19.m02115; chaperone clpB
Length=833
Score = 114 bits (286), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 104/434 (23%), Positives = 196/434 (45%), Gaps = 89/434 (20%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DEIH LI ++ + +LK + + +IG+TT EY Y +D AF RRF+ +
Sbjct 272 DEIHHLIQ--NQENGVNVTNLLKPIMTSTLVKIIGSTTAKEYHQYFRRDRAFERRFEILR 329
Query 78 VEAPSKERTLSILRKVKGNYEAHHSLQIPDLLLSQIVQLSDQLLKRRAFPDKALDLLDEA 137
+ S + TL+IL + + E +H ++I D L V+LS + + R PDKA+DLLDEA
Sbjct 330 LHENSADETLAILHGSRPSLEDYHGVKITDDALVASVELSTRFIPNRYLPDKAIDLLDEA 389
Query 138 ---------CA---FRRVRHNSRRAELALLLQQQRDGKTHFAEEELAALQAEY------- 178
C+ F +RH + ALL + + + + E+ ++++ Y
Sbjct 390 AMLSKRKRSCSNDLFELLRHKTGIIS-ALLRGKSGNDQLNRVLSEIESMESRYKSLHGKL 448
Query 179 -------------AEMTEPTEERPD--PEVLELQM---DDVAHVLS-------------- 206
+T+ +E + PE++ L + D+ V++
Sbjct 449 RDVQQQQQNHEKNGNLTKASELKNHKIPEIIRLIVQLQSDIKMVINKTIEDSKDMLPVDM 508
Query 207 ---------------LWTGIPVGRMTEDEA---TRVLKLADLLSKRVIGQDEAVQAVANA 248
+++G+ V R+ A + + ++ +L ++ ++AVQ ++N
Sbjct 509 LNDGNTGVSPASDPDVFSGVYVDRIDIASALSQSTGIPVSVVLRSQMGHYEQAVQQLSNI 568
Query 249 --------------IRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNL 294
I + + ++ +K A ++G GVGK L ++ + K +
Sbjct 569 VLGQNEAVYKTLLHIYMYSSSIASGRKIGAALYYVGPPGVGKRLLLNHVSRMFGMALKYI 628
Query 295 IRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSL 354
++V +++ + L+G PPGY+G+ EGG L E ++ P+ +V F++ H N+ +L
Sbjct 629 CGSNLV---SSNATNILVGSPPGYIGHREGGMLCEWIKDHPYGIVAFEDAHLLHVNVVNL 685
Query 355 LLPMLDEGHLTDTK 368
L+ +D G L D +
Sbjct 686 LIGAIDNGFLVDNQ 699
> tpv:TP05_0023 clpC; molecular chaperone
Length=502
Score = 112 bits (279), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 57/143 (39%), Positives = 85/143 (59%), Gaps = 5/143 (3%)
Query 235 VIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNL 294
+ GQ+E + ++ + R ++ KPI FL G SG GKT++ K L+ + ++NL
Sbjct 232 IFGQNENIDIISKKLI--RLDNNKYNKPIANFLLCGPSGTGKTDICKILSTYLGSEKQNL 289
Query 295 IRLDMVEYQEAHSISRLIGPPPGYMG---NDEGGQLTEAVRQKPHSVVLFDEVENAHKNL 351
I+LDM E E HS+SRL+G PPGY+G + + L + + KP+SVVL DE+E A+K L
Sbjct 290 IKLDMSELAEEHSVSRLLGSPPGYVGGGKSKKSKTLVDEIIDKPNSVVLLDEIEKAYKRL 349
Query 352 WSLLLPMLDEGHLTDTKQRAGGL 374
+ L +LDEG L D+ G
Sbjct 350 CYIFLQILDEGILIDSSGTVGNF 372
> tgo:TGME49_102000 chaperone clpB protein, putative
Length=240
Score = 89.4 bits (220), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 38/69 (55%), Positives = 55/69 (79%), Gaps = 0/69 (0%)
Query 299 MVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSLLLPM 358
M E+ E HS+S+LIG PPGY+G + G LTEA+ +KP +V+LFDE+E AHK++ +L+L +
Sbjct 1 MSEFMEKHSLSKLIGAPPGYVGYGKSGLLTEAIAKKPFTVLLFDEIEKAHKDINNLMLQL 60
Query 359 LDEGHLTDT 367
LD+G LTD+
Sbjct 61 LDDGKLTDS 69
> ath:AT3G45450 Clp amino terminal domain-containing protein
Length=341
Score = 52.4 bits (124), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 41/72 (56%), Gaps = 9/72 (12%)
Query 18 DEIHTLIGAGKAAGSMDASQMLKVPLARGEIVLIGATTLSEYKLYLEKDAAFCRRFQKIL 77
DE+H LIGAG G++DA+ +LK L R E+ +Y+ ++E D A RRFQ +
Sbjct 247 DEMHLLIGAGAVEGAIDAANILKPALERCEL---------QYRKHIENDPALERRFQPVK 297
Query 78 VEAPSKERTLSI 89
V P+ E + I
Sbjct 298 VPEPTVEEAIQI 309
> cpv:cgd7_2620 ClpB ATpase (bacterial), signal peptide
Length=1263
Score = 51.2 bits (121), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 47/189 (24%), Positives = 87/189 (46%), Gaps = 37/189 (19%)
Query 195 ELQMDDVAHVLSLWTGIPVGRMTEDEATRVL------KLADLLSKRVIGQDEAVQAVANA 248
E+ +A ++S G + ++ ++ R L +L+++LSK VIGQ A+ V+
Sbjct 908 EIDASHIAFIISERYGKSITKLLDEIEYRKLPERMKDRLSEILSKYVIGQKSAIDYVS-- 965
Query 249 IRNHRAG---LSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDM------ 299
G L ++K LF+G SG GK A AL + +E +++ D
Sbjct 966 ---FHLGVELLRDNKNNPRCLLFVGPSGSGKKTFALALQTAL--TESSVLFYDFSYDITM 1020
Query 300 ---------VEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKN 350
++ +A + RL+G P + G ++E ++Q +V F+ +EN H +
Sbjct 1021 YSHFKILKSSDFADADAAKRLLGDNP------KTGIISEELKQTGKTVFFFEHIENMHPD 1074
Query 351 LWSLLLPML 359
+ L+L +L
Sbjct 1075 VIKLILDIL 1083
> ath:AT1G07200 ATP-dependent Clp protease ClpB protein-related
Length=979
Score = 47.8 bits (112), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 59/122 (48%), Gaps = 9/122 (7%)
Query 227 LADLLSKRVIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFL-FLGSSGVGKTELAKALAE 285
L ++LS++V Q EAV A++ I + + + G +L LG VGK ++A L+E
Sbjct 621 LREILSRKVAWQTEAVNAISQIICGCKTDSTRRNQASGIWLALLGPDKVGKKKVAMTLSE 680
Query 286 EIFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVE 345
F + N I +D E S+ + G +T + +KPHSVVL + VE
Sbjct 681 VFFGGKVNYICVDF--GAEHCSLDD------KFRGKTVVDYVTGELSRKPHSVVLLENVE 732
Query 346 NA 347
A
Sbjct 733 KA 734
> pfa:PFI0355c ATP-dependent heat shock protein, putative; K03667
ATP-dependent HslUV protease ATP-binding subunit HslU
Length=922
Score = 45.8 bits (107), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/63 (42%), Positives = 41/63 (65%), Gaps = 5/63 (7%)
Query 227 LADLLSKRVIGQDEAVQAVANAIRN--HRAGLSED-KKPI--GAFLFLGSSGVGKTELAK 281
+ + L+K +IGQ EA + VANA+R R +S+D KK I L +G +GVGKTE+A+
Sbjct 490 IVEYLNKYIIGQSEAKKVVANALRQRWRRIQVSDDMKKDIIPKNILMIGPTGVGKTEIAR 549
Query 282 ALA 284
++
Sbjct 550 RIS 552
> ath:AT5G45720 ATP binding / DNA binding / DNA-directed DNA polymerase/
nucleoside-triphosphatase/ nucleotide binding
Length=966
Score = 45.8 bits (107), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 34/134 (25%), Positives = 61/134 (45%), Gaps = 14/134 (10%)
Query 235 VIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEI-FHSEKN 293
++GQ+ VQA++NAI R GL ++F G +G GKT A+ A + HS +
Sbjct 357 LLGQNLVVQALSNAIAKRRVGL--------LYVFHGPNGTGKTSCARVFARALNCHSTEQ 408
Query 294 L----IRLDMVEYQEAHS-ISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAH 348
+ V Y + + R +GP + + + +QK V++FD+ +
Sbjct 409 SKPCGVCSSCVSYDDGKNRYIREMGPVKSFDFENLLDKTNIRQQQKQQLVLIFDDCDTMS 468
Query 349 KNLWSLLLPMLDEG 362
+ W+ L ++D
Sbjct 469 TDCWNTLSKIVDRA 482
> eco:b3931 hslU, clpY, ECK3923, htpI, JW3902; molecular chaperone
and ATPase component of HslUV protease; K03667 ATP-dependent
HslUV protease ATP-binding subunit HslU
Length=443
Score = 42.0 bits (97), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 26/60 (43%), Positives = 38/60 (63%), Gaps = 5/60 (8%)
Query 231 LSKRVIGQDEAVQAVANAIRN--HRAGLSEDKK---PIGAFLFLGSSGVGKTELAKALAE 285
L K +IGQD A ++VA A+RN R L+E+ + L +G +GVGKTE+A+ LA+
Sbjct 13 LDKHIIGQDNAKRSVAIALRNRWRRMQLNEELRHEVTPKNILMIGPTGVGKTEIARRLAK 72
> tgo:TGME49_089780 ATP-dependent heat shock protein, putative
(EC:2.7.1.71); K03667 ATP-dependent HslUV protease ATP-binding
subunit HslU
Length=567
Score = 39.3 bits (90), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 47/86 (54%), Gaps = 11/86 (12%)
Query 227 LADLLSKRVIGQDEAVQAVANAIRNHR-------AGLSEDKKPIGAFLFLGSSGVGKTEL 279
+ + L++ ++GQ EA +AVA A+R L ED P L +G +GVGKTE+
Sbjct 1 MVEYLNRFIVGQVEAKRAVAVALRQRWRRRHIEDERLREDITPKN-ILLIGPTGVGKTEV 59
Query 280 AKALAEEIFHSEKNLIRLDMVEYQEA 305
A+ LA+ + + I+++ ++ E
Sbjct 60 ARRLAKRL---DAPFIKVEATKFTEV 82
> ath:AT5G40090 ATP binding / nucleoside-triphosphatase/ nucleotide
binding / transmembrane receptor
Length=459
Score = 39.3 bits (90), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 29/102 (28%), Positives = 54/102 (52%), Gaps = 7/102 (6%)
Query 256 LSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIG-- 313
++++ + IG + GS+GVGKT LA+ + EIF + + + LD VE + + + G
Sbjct 199 VNKEVRTIGIW---GSAGVGKTTLARYIYAEIFVNFQTHVFLDNVENMK-DKLLKFEGEE 254
Query 314 -PPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSL 354
P +G ++TEA R+ +++ D+V N + W +
Sbjct 255 DPTVIISSYHDGHEITEARRKHRKILLIADDVNNMEQGKWII 296
> ath:AT4G30350 heat shock protein-related
Length=924
Score = 38.9 bits (89), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 40/149 (26%), Positives = 69/149 (46%), Gaps = 10/149 (6%)
Query 226 KLADLLSKRVIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAE 285
KL L+K V Q +A +VA AI + G + K I +F G GK+++A AL++
Sbjct 572 KLLKGLAKSVWWQHDAASSVAAAITECKHGNGKSKGDIW-LMFTGPDRAGKSKMASALSD 630
Query 286 EIFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVE 345
+ S+ I L S SR+ G + EAVR+ P +V++ ++++
Sbjct 631 LVSGSQPITISL--------GSSSRM-DDGLNIRGKTALDRFAEAVRRNPFAVIVLEDID 681
Query 346 NAHKNLWSLLLPMLDEGHLTDTKQRAGGL 374
A L + + ++ G + D+ R L
Sbjct 682 EADILLRNNVKIAIERGRICDSYGREVSL 710
> ath:AT4G18820 ATP binding / DNA binding / DNA-directed DNA polymerase/
nucleoside-triphosphatase/ nucleotide binding
Length=1111
Score = 38.9 bits (89), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 34/137 (24%), Positives = 60/137 (43%), Gaps = 22/137 (16%)
Query 235 VIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEI-FHSEKN 293
++GQ+ VQA++NA+ + GL ++F G +G GKT A+ A + HS +
Sbjct 445 LLGQNLVVQALSNAVARRKLGL--------LYVFHGPNGTGKTSCARIFARALNCHSMEQ 496
Query 294 LIRLDMVEYQEAHSIS-----RLIGPPPGY-----MGNDEGGQLTEAVRQKPHSVVLFDE 343
+H + R +GP Y M +G + + Q P V +FD+
Sbjct 497 PKPCGTCSSCVSHDMGKSWNIREVGPVGNYDFEKIMDLLDGNVMVSS--QSPR-VFIFDD 553
Query 344 VENAHKNLWSLLLPMLD 360
+ + W+ L ++D
Sbjct 554 CDTLSSDCWNALSKVVD 570
> tgo:TGME49_010730 ATP-dependent protease ATP-binding subunit,
putative (EC:3.1.3.48)
Length=565
Score = 37.0 bits (84), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 25/60 (41%), Positives = 37/60 (61%), Gaps = 6/60 (10%)
Query 231 LSKRVIGQDEAVQAVANAIRNH--RAGLSEDK--KPIGA--FLFLGSSGVGKTELAKALA 284
L K ++GQD A +++A A+R+ R + ++K + I L +G SG GKTELAK LA
Sbjct 80 LDKYIVGQDTAKKSLAIALRDRWRRQQVKDEKLRREIAPNNLLLIGPSGCGKTELAKRLA 139
> ath:AT5G46260 disease resistance protein (TIR-NBS-LRR class),
putative
Length=1205
Score = 36.2 bits (82), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 33/116 (28%), Positives = 56/116 (48%), Gaps = 6/116 (5%)
Query 177 EYAEMTEPTEERPDPEVLELQMDDVAHVLSLWTGIPVGRMTEDEATRVLKLA-DLLSKRV 235
E+ ++ E T +R EV + HV ++ G + +DEA + ++A D+L K +
Sbjct 115 EFGKIFEKTCKRQTEEVKNQWKKALTHVANM-LGFDSSKW-DDEAKMIEEIANDVLRKLL 172
Query 236 IGQD---EAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIF 288
+ E + + I N A L + K + GSSG+GKT +A+AL +F
Sbjct 173 LTTSKDFEDFVGLEDHIANMSALLDLESKEVKMVGIWGSSGIGKTTIARALFNNLF 228
> bbo:BBOV_II000190 18.m05996; ATPase, AAA family; K12196 vacuolar
protein-sorting-associated protein 4
Length=363
Score = 35.8 bits (81), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 23/86 (26%), Positives = 39/86 (45%), Gaps = 13/86 (15%)
Query 261 KPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVEYQEAHSISRLIGPPPGYMG 320
KP L G G GKT LAKA A E+ S + + +S+ +G ++
Sbjct 147 KPWRGILLYGPPGTGKTYLAKACATELDAS--------FIAISSSDVLSKWLGESEKFVK 198
Query 321 NDEGGQLTEAVRQKPHSVVLFDEVEN 346
+ L +A R++ V+ DE+++
Sbjct 199 S-----LFQAARERAPCVIFIDEIDS 219
> ath:AT2G27600 SKD1; SKD1 (SUPPRESSOR OF K+ TRANSPORT GROWTH
DEFECT1); ATP binding / nucleoside-triphosphatase/ nucleotide
binding; K12196 vacuolar protein-sorting-associated protein
4
Length=435
Score = 35.4 bits (80), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 31/117 (26%), Positives = 50/117 (42%), Gaps = 20/117 (17%)
Query 235 VIGQDEAVQAVANAI---RNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIFHSE 291
V G + A QA+ A+ + ++P AFL G G GK+ LAKA+A E +
Sbjct 134 VAGLESAKQALQEAVILPVKFPQFFTGKRRPWRAFLLYGPPGTGKSYLAKAVATEADSTF 193
Query 292 KNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEG--GQLTEAVRQKPHSVVLFDEVEN 346
++ D+V +MG E L E R+ S++ DE+++
Sbjct 194 FSVSSSDLVS---------------KWMGESEKLVSNLFEMARESAPSIIFVDEIDS 235
> ath:AT1G62130 AAA-type ATPase family protein
Length=1025
Score = 35.4 bits (80), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 23/42 (54%), Gaps = 3/42 (7%)
Query 261 KPIGAFLFLGSSGVGKTELAKALAEEIFHSEKNLIRLDMVEY 302
KP L G SG GKT LAKA+A E + NLI + M +
Sbjct 768 KPCNGILLFGPSGTGKTMLAKAVATE---AGANLINMSMSRW 806
> dre:565062 im:7165310; si:ch73-178d14.1
Length=336
Score = 35.0 bits (79), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 35/138 (25%), Positives = 56/138 (40%), Gaps = 22/138 (15%)
Query 231 LSKRVIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFL--GSSGVGKTELAKALAEEIF 288
L ++ GQ A Q + A+ G +KKP + G +G GK +++ LAE I+
Sbjct 71 LDTKLYGQHVAGQVILKAV----TGFMNNKKPKKPLVLSLHGWTGTGKNFVSQLLAENIY 126
Query 289 HS--EKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQK----PHSVVLFD 342
E + + L H I + QL + +R P S+ +FD
Sbjct 127 VKGMESSFVHLFTATAHFPHEIHI----------DTYKTQLQDWIRGNVSICPRSMFIFD 176
Query 343 EVENAHKNLWSLLLPMLD 360
E++ H L + P LD
Sbjct 177 EMDKMHPGLIDSIKPYLD 194
> dre:100170794 zgc:194342
Length=323
Score = 35.0 bits (79), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 33/143 (23%), Positives = 57/143 (39%), Gaps = 20/143 (13%)
Query 231 LSKRVIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFL--FLGSSGVGKTELAKALAEEIF 288
K + GQ V++ + + DK P + F G++G GK +AK +A ++
Sbjct 55 FDKSLYGQ----HIVSDVVPKSVSFFMTDKNPNKPLVLSFHGTAGTGKNHVAKIIARNVY 110
Query 289 HS-EKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEA----VRQKPHSVVLFDE 343
EK+ + H+ P + QL + V P S+ +FDE
Sbjct 111 KKGEKS---------KHVHTFISQFHFPHQENVHMYSAQLKQWIHGNVSSFPRSMFIFDE 161
Query 344 VENAHKNLWSLLLPMLDEGHLTD 366
++ H L ++ P LD + D
Sbjct 162 MDKMHPELIDIIKPFLDYNYNVD 184
> mmu:30931 Tor1a, DQ2, Dyt1, MGC18883, torsinA; torsin family
1, member A (torsin A)
Length=333
Score = 35.0 bits (79), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 33/140 (23%), Positives = 64/140 (45%), Gaps = 14/140 (10%)
Query 231 LSKRVIGQDEAVQAVANAIRNHRAGLSEDKKPIGAFLFLGSSGVGKTELAKALAEEIF-- 288
L ++ GQ A + + NA+ + + KKP+ L G +G GK +K +AE I+
Sbjct 66 LDNKLFGQHLAKKVILNAVSGFLSN-PKPKKPLTLSLH-GWTGTGKNFASKIIAENIYEG 123
Query 289 --HSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVEN 346
+S+ + + + + A +I++ ++ G ++ R S+ +FDE++
Sbjct 124 GLNSDYVHLFVATLHFPHASNITQYKDQLQMWIR----GNVSACAR----SIFIFDEMDK 175
Query 347 AHKNLWSLLLPMLDEGHLTD 366
H L + P LD + D
Sbjct 176 MHAGLIDAIKPFLDYYDVVD 195
Lambda K H
0.316 0.133 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 20093762788
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40