bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
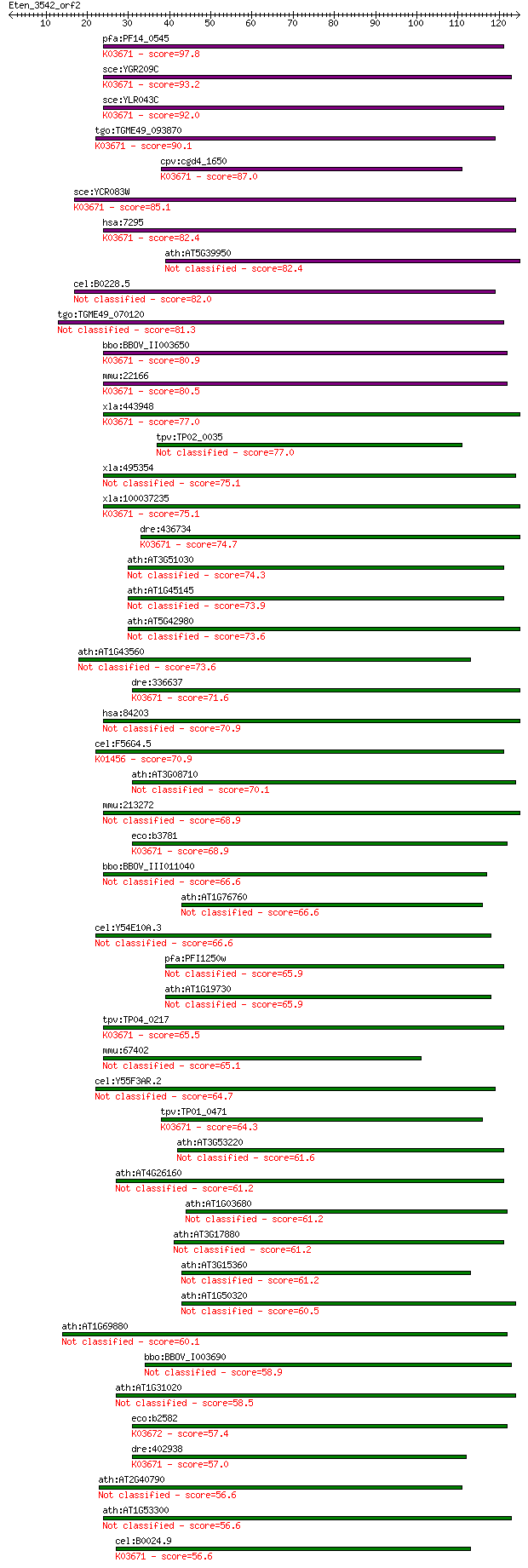
Query= Eten_3542_orf2
Length=124
Score E
Sequences producing significant alignments: (Bits) Value
pfa:PF14_0545 trx; thioredoxin, putative; K03671 thioredoxin 1 97.8
sce:YGR209C TRX2, LMA1; Cytoplasmic thioredoxin isoenzyme of t... 93.2 2e-19
sce:YLR043C TRX1, LMA1; Cytoplasmic thioredoxin isoenzyme of t... 92.0 4e-19
tgo:TGME49_093870 thioredoxin, putative (EC:5.3.4.1); K03671 t... 90.1 1e-18
cpv:cgd4_1650 possible thioredoxin H-type of possible fungal o... 87.0 1e-17
sce:YCR083W TRX3; Mitochondrial thioredoxin, highly conserved ... 85.1 5e-17
hsa:7295 TXN, DKFZp686B1993, MGC61975, TRX, TRX1; thioredoxin;... 82.4 3e-16
ath:AT5G39950 ATTRX2; ATTRX2 (THIOREDOXIN 2); oxidoreductase, ... 82.4 3e-16
cel:B0228.5 trx-1; ThioRedoXin [see also xtr] family member (t... 82.0 4e-16
tgo:TGME49_070120 thioredoxin, putative (EC:5.3.4.1) 81.3 7e-16
bbo:BBOV_II003650 18.m06306; thioredoxin; K03671 thioredoxin 1 80.9
mmu:22166 Txn1, ADF, AW550880, Trx1, Txn; thioredoxin 1; K0367... 80.5 1e-15
xla:443948 MGC80314 protein; K03671 thioredoxin 1 77.0
tpv:TP02_0035 thioredoxin 77.0 1e-14
xla:495354 txn, trx; thioredoxin 75.1 5e-14
xla:100037235 hypothetical protein LOC100037235; K03671 thiore... 75.1 5e-14
dre:436734 zgc:92903; K03671 thioredoxin 1 74.7
ath:AT3G51030 ATTRX1; ATTRX1; oxidoreductase, acting on sulfur... 74.3 8e-14
ath:AT1G45145 ATTRX5; ATTRX5; oxidoreductase, acting on sulfur... 73.9 1e-13
ath:AT5G42980 ATTRX3; ATTRX3 (THIOREDOXIN 3); oxidoreductase, ... 73.6 1e-13
ath:AT1G43560 Aty2; Aty2 (Arabidopsis thioredoxin y2); electro... 73.6 1e-13
dre:336637 TXN, fb15h09, wu:fb15h09; zgc:56493; K03671 thiored... 71.6 6e-13
hsa:84203 TXNDC2, DKFZp434H0311, MGC35026, SPTRX, SPTRX1; thio... 70.9 1e-12
cel:F56G4.5 png-1; PNG (Peptide:N-Glycanase) homolog family me... 70.9 1e-12
ath:AT3G08710 ATH9; ATH9 (thioredoxin H-type 9) 70.1 2e-12
mmu:213272 Txndc2, AU021712, Sptrx-1, Sptrx1, Trx4; thioredoxi... 68.9 3e-12
eco:b3781 trxA, dasC, ECK3773, fipA, JW5856, tsnC; thioredoxin... 68.9 4e-12
bbo:BBOV_III011040 17.m07951; thioredoxin domain containing pr... 66.6 2e-11
ath:AT1G76760 ATY1; ATY1 (ARABIDOPSIS THIOREDOXIN Y1); electro... 66.6 2e-11
cel:Y54E10A.3 hypothetical protein 66.6 2e-11
pfa:PFI1250w tlp2; Tlp2 65.9 3e-11
ath:AT1G19730 ATTRX4; ATTRX4; oxidoreductase, acting on sulfur... 65.9 3e-11
tpv:TP04_0217 thioredoxin; K03671 thioredoxin 1 65.5
mmu:67402 Txndc8, 4930429J24Rik, MGC151140, MGC151142, Sptrx-3... 65.1 6e-11
cel:Y55F3AR.2 hypothetical protein 64.7 6e-11
tpv:TP01_0471 thioredoxin; K03671 thioredoxin 1 64.3
ath:AT3G53220 thioredoxin family protein 61.6 6e-10
ath:AT4G26160 ACHT1; ACHT1 (ATYPICAL CYS HIS RICH THIOREDOXIN ... 61.2 8e-10
ath:AT1G03680 ATHM1; ATHM1; enzyme activator 61.2 8e-10
ath:AT3G17880 ATTDX; ATTDX (TETRATICOPEPTIDE DOMAIN-CONTAINING... 61.2 9e-10
ath:AT3G15360 TRX-M4; TRX-M4 (ARABIDOPSIS THIOREDOXIN M-TYPE 4... 61.2 9e-10
ath:AT1G50320 ATHX; ATHX; enzyme activator 60.5 1e-09
ath:AT1G69880 ATH8; ATH8 (thioredoxin H-type 8) 60.1 2e-09
bbo:BBOV_I003690 19.m02235; thioredoxin 58.9 4e-09
ath:AT1G31020 ATO2; ATO2 (Arabidopsis thioredoxin o2) 58.5 5e-09
eco:b2582 trxC, ECK2580, JW2566, yfiG; thioredoxin 2 (EC:1.8.1... 57.4 1e-08
dre:402938 txn2, MGC77127, zgc:77127; thioredoxin 2; K03671 th... 57.0 2e-08
ath:AT2G40790 ATCXXS2 (C-TERMINAL CYSTEINE RESIDUE IS CHANGED ... 56.6 2e-08
ath:AT1G53300 TTL1; TTL1 (TETRATRICOPETIDE-REPEAT THIOREDOXIN-... 56.6 2e-08
cel:B0024.9 trx-2; ThioRedoXin [see also xtr] family member (t... 56.6 2e-08
> pfa:PF14_0545 trx; thioredoxin, putative; K03671 thioredoxin
1
Length=104
Score = 97.8 bits (242), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 49/98 (50%), Positives = 64/98 (65%), Gaps = 1/98 (1%)
Query 24 VKHLKGIEEFDEAI-KNGLAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDENP 82
VK + EFD I +N L IVDFFA WCGPCK I PF EE S T T + F VDVDE
Sbjct 2 VKIVTSQAEFDSIISQNELVIVDFFAEWCGPCKRIAPFYEECSKTYTKMVFIKVDVDEVS 61
Query 83 ELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEAL 120
E+ E+E + +MPTFK+YK+G V+T++G L++ +
Sbjct 62 EVTEKENITSMPTFKVYKNGSSVDTLLGANDSALKQLI 99
> sce:YGR209C TRX2, LMA1; Cytoplasmic thioredoxin isoenzyme of
the thioredoxin system which protects cells against oxidative
and reductive stress, forms LMA1 complex with Pbi2p, acts
as a cofactor for Tsa1p, required for ER-Golgi transport and
vacuole inheritance; K03671 thioredoxin 1
Length=104
Score = 93.2 bits (230), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 42/101 (41%), Positives = 63/101 (62%), Gaps = 2/101 (1%)
Query 24 VKHLKGIEEFDEAIKNG--LAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDEN 81
V LK E+D A+ +G L +VDFFATWCGPCKMI P +E+ + + FY +DVDE
Sbjct 2 VTQLKSASEYDSALASGDKLVVVDFFATWCGPCKMIAPMIEKFAEQYSDAAFYKLDVDEV 61
Query 82 PELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEALNN 122
++ ++ +V +MPT YK GK V +VG +++A+ +
Sbjct 62 SDVAQKAEVSSMPTLIFYKGGKEVTRVVGANPAAIKQAIAS 102
> sce:YLR043C TRX1, LMA1; Cytoplasmic thioredoxin isoenzyme of
the thioredoxin system which protects cells against oxidative
and reductive stress, forms LMA1 complex with Pbi2p, acts
as a cofactor for Tsa1p, required for ER-Golgi transport and
vacuole inheritance; K03671 thioredoxin 1
Length=103
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 43/98 (43%), Positives = 63/98 (64%), Gaps = 1/98 (1%)
Query 24 VKHLKGIEEFDEAI-KNGLAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDENP 82
V K EFD AI ++ L +VDF+ATWCGPCKMI P +E+ S +FY +DVDE
Sbjct 2 VTQFKTASEFDSAIAQDKLVVVDFYATWCGPCKMIAPMIEKFSEQYPQADFYKLDVDELG 61
Query 83 ELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEAL 120
++ ++ +V AMPT L+K+GK V +VG +++A+
Sbjct 62 DVAQKNEVSAMPTLLLFKNGKEVAKVVGANPAAIKQAI 99
> tgo:TGME49_093870 thioredoxin, putative (EC:5.3.4.1); K03671
thioredoxin 1
Length=106
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 42/100 (42%), Positives = 63/100 (63%), Gaps = 3/100 (3%)
Query 22 MPVKHLKGIEEFDEAIK-NGLAIVDFFATWCGPCKMIRPFLEEKSNT--MTHINFYGVDV 78
MPV H+ +F I+ N + +VDF+A WCGPC+ + P +E S + F +DV
Sbjct 1 MPVHHVTTEAQFKSLIEENEMVLVDFYAVWCGPCRQVAPLVEAMSEKPEYAKVKFVKIDV 60
Query 79 DENPELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEE 118
DE ++ ERE++ AMPTFKL+K GK V+T++G ++EE
Sbjct 61 DELADVAEREEINAMPTFKLFKQGKAVDTVLGANAERVEE 100
> cpv:cgd4_1650 possible thioredoxin H-type of possible fungal
or plant origin, small protein ; K03671 thioredoxin 1
Length=105
Score = 87.0 bits (214), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 38/73 (52%), Positives = 51/73 (69%), Gaps = 0/73 (0%)
Query 38 KNGLAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDENPELCEREKVQAMPTFK 97
+ GL +VD+FATWCGPC MI P LEE S + + F VDVD P + + E V+AMPTFK
Sbjct 18 RQGLVVVDYFATWCGPCNMIAPKLEEFSEQVKDVCFIKVDVDSLPSIADAESVRAMPTFK 77
Query 98 LYKDGKVVETIVG 110
+ +GK V+++VG
Sbjct 78 FFLNGKEVKSVVG 90
> sce:YCR083W TRX3; Mitochondrial thioredoxin, highly conserved
oxidoreductase required to maintain the redox homeostasis
of the cell, forms the mitochondrial thioredoxin system with
Trr2p, redox state is maintained by both Trr2p and Glr1p; K03671
thioredoxin 1
Length=127
Score = 85.1 bits (209), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 41/108 (37%), Positives = 64/108 (59%), Gaps = 1/108 (0%)
Query 17 FRLAKMPVKHLKGIEEFDEAIK-NGLAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYG 75
F+ + + L + EF IK N ++DF+ATWCGPCKM++P L + + F
Sbjct 20 FQSSYTSITKLTNLTEFRNLIKQNDKLVIDFYATWCGPCKMMQPHLTKLIQAYPDVRFVK 79
Query 76 VDVDENPELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEALNNL 123
DVDE+P++ + +V AMPTF L KDG+++ I+G LE+ + +L
Sbjct 80 CDVDESPDIAKECEVTAMPTFVLGKDGQLIGKIIGANPTALEKGIKDL 127
> hsa:7295 TXN, DKFZp686B1993, MGC61975, TRX, TRX1; thioredoxin;
K03671 thioredoxin 1
Length=105
Score = 82.4 bits (202), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 42/103 (40%), Positives = 60/103 (58%), Gaps = 3/103 (2%)
Query 24 VKHLKGIEEFDEAIK---NGLAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDE 80
VK ++ F EA+ + L +VDF ATWCGPCKMI+PF S +++ F VDVD+
Sbjct 2 VKQIESKTAFQEALDAAGDKLVVVDFSATWCGPCKMIKPFFHSLSEKYSNVIFLEVDVDD 61
Query 81 NPELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEALNNL 123
++ +V+ MPTF+ +K G+ V G K KLE +N L
Sbjct 62 CQDVASECEVKCMPTFQFFKKGQKVGEFSGANKEKLEATINEL 104
> ath:AT5G39950 ATTRX2; ATTRX2 (THIOREDOXIN 2); oxidoreductase,
acting on sulfur group of donors, disulfide as acceptor
Length=133
Score = 82.4 bits (202), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 37/86 (43%), Positives = 56/86 (65%), Gaps = 0/86 (0%)
Query 39 NGLAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDENPELCEREKVQAMPTFKL 98
N L +VDF A+WCGPC+MI P + ++ ++F +DVDE P++ + V AMPTF L
Sbjct 47 NKLLVVDFSASWCGPCRMIEPAIHAMADKFNDVDFVKLDVDELPDVAKEFNVTAMPTFVL 106
Query 99 YKDGKVVETIVGVLKPKLEEALNNLK 124
K GK +E I+G K +LE+ ++ L+
Sbjct 107 VKRGKEIERIIGAKKDELEKKVSKLR 132
> cel:B0228.5 trx-1; ThioRedoXin [see also xtr] family member
(trx-1)
Length=114
Score = 82.0 bits (201), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 40/105 (38%), Positives = 61/105 (58%), Gaps = 3/105 (2%)
Query 17 FRLAKMPVKHLKGIEEFDEAIKNG---LAIVDFFATWCGPCKMIRPFLEEKSNTMTHINF 73
L K P+ L + +F++ I+ + I+DF+ATWCGPCK I P +E + T I F
Sbjct 1 MSLTKEPILELADMSDFEQLIRQHPEKIIILDFYATWCGPCKAIAPLYKELATTHKGIIF 60
Query 74 YGVDVDENPELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEE 118
VDVDE +LC + V+ MPTF K+G +E + G ++ +L +
Sbjct 61 CKVDVDEAEDLCSKYDVKMMPTFIFTKNGDAIEALEGCVEDELRQ 105
> tgo:TGME49_070120 thioredoxin, putative (EC:5.3.4.1)
Length=178
Score = 81.3 bits (199), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 42/114 (36%), Positives = 67/114 (58%), Gaps = 7/114 (6%)
Query 13 FIFDFRLAKMPVKHLKGIEEFDEAIKN----GLAIVDFFATWCGPCKMIRPFLEEKSNTM 68
F DF++ + V +K E+F + + + +V F A+WCGPC+ + P +E S M
Sbjct 61 FCGDFQV-RTIVNRVKKAEDFQKVTDSEEDCSVKVVQFSASWCGPCRQVTPTIEGWSEKM 119
Query 69 --THINFYGVDVDENPELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEAL 120
+ + F+ VD+DE PEL E + ++PTF +K+GK V T+VG K+EEA+
Sbjct 120 PASEVQFFHVDIDECPELAEEYDISSVPTFLFFKNGKKVNTVVGGNTAKIEEAI 173
> bbo:BBOV_II003650 18.m06306; thioredoxin; K03671 thioredoxin
1
Length=103
Score = 80.9 bits (198), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 38/99 (38%), Positives = 58/99 (58%), Gaps = 1/99 (1%)
Query 24 VKHLKGIEEFDEAIKN-GLAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDENP 82
VK + ++EF ++ GL +VDFFATWCGPC P E + + + F VD+ E P
Sbjct 2 VKQIASMDEFYSILQTPGLVVVDFFATWCGPCMNFAPKFENFAREYSSVTFVKVDISEFP 61
Query 83 ELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEALN 121
EL + + ++P FKL+K+G VV +VG L+ A++
Sbjct 62 ELQTKYAITSIPAFKLFKNGDVVGEVVGASAINLKNAID 100
> mmu:22166 Txn1, ADF, AW550880, Trx1, Txn; thioredoxin 1; K03671
thioredoxin 1
Length=105
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 41/101 (40%), Positives = 60/101 (59%), Gaps = 3/101 (2%)
Query 24 VKHLKGIEEFDEAIK---NGLAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDE 80
VK ++ E F EA+ + L +VDF ATWCGPCKMI+PF + +++ F VDVD+
Sbjct 2 VKLIESKEAFQEALAAAGDKLVVVDFSATWCGPCKMIKPFFHSLCDKYSNVVFLEVDVDD 61
Query 81 NPELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEALN 121
++ +V+ MPTF+ YK G+ V G K KLE ++
Sbjct 62 CQDVAADCEVKCMPTFQFYKKGQKVGEFSGANKEKLEASIT 102
> xla:443948 MGC80314 protein; K03671 thioredoxin 1
Length=105
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 41/104 (39%), Positives = 58/104 (55%), Gaps = 3/104 (2%)
Query 24 VKHLKGIEEFDEAIKNG---LAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDE 80
V+H++ +EEF +K L +VDF ATWCGPCKMI P E+ S F VDVD+
Sbjct 2 VRHIENLEEFQLVLKEAGGKLVVVDFTATWCGPCKMIAPVFEKLSVDNPDAVFLKVDVDD 61
Query 81 NPELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEALNNLK 124
++ V+ MPTF+ YK+G V+ G + L + + LK
Sbjct 62 AQDVAAHCDVKCMPTFQFYKNGIKVDEFSGANQSSLIQKVEALK 105
> tpv:TP02_0035 thioredoxin
Length=133
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 31/74 (41%), Positives = 51/74 (68%), Gaps = 0/74 (0%)
Query 37 IKNGLAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDENPELCEREKVQAMPTF 96
+++ L + F A+WC PC+ +P +EE SN + H+NF VDVD P++ + E V+ +P F
Sbjct 38 LQDKLVLAKFSASWCKPCQKAKPVVEELSNELPHLNFIDVDVDNLPQIADEEGVKTIPFF 97
Query 97 KLYKDGKVVETIVG 110
KL+K+GK+++ I G
Sbjct 98 KLFKNGKLLDEITG 111
> xla:495354 txn, trx; thioredoxin
Length=105
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 39/103 (37%), Positives = 58/103 (56%), Gaps = 3/103 (2%)
Query 24 VKHLKGIEEFDEAIKNG---LAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDE 80
V++L +EE+ A+K+ L ++DF A WCGPC+ I P E+ S I + VDVD
Sbjct 2 VRYLNSLEEYHCALKDAGEKLVVIDFTAVWCGPCQRIAPDFEKLSTENPDIVLFKVDVDN 61
Query 81 NPELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEALNNL 123
++ + V++MPTF YK GK VE G KL+ ++ L
Sbjct 62 ASDVAQLCGVRSMPTFVFYKSGKEVERFSGADISKLKSTISRL 104
> xla:100037235 hypothetical protein LOC100037235; K03671 thioredoxin
1
Length=105
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 41/104 (39%), Positives = 58/104 (55%), Gaps = 3/104 (2%)
Query 24 VKHLKGIEEFDEAI---KNGLAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDE 80
V+H++ +EEF + K L +VDF ATWCGPCKMI P E+ S + F VDVD+
Sbjct 2 VRHVETLEEFQNVLQEAKEKLVVVDFTATWCGPCKMIAPVFEKLSVENPDVVFLKVDVDD 61
Query 81 NPELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEALNNLK 124
++ +V+ MPTF YK+G V G + L + + LK
Sbjct 62 AQDVAAHCEVKCMPTFHFYKNGLKVFEFSGANESSLVQKVAELK 105
> dre:436734 zgc:92903; K03671 thioredoxin 1
Length=107
Score = 74.7 bits (182), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 42/98 (42%), Positives = 57/98 (58%), Gaps = 7/98 (7%)
Query 33 FDEAIKNG---LAIVDFFATWCGPCKMIRPF---LEEKSNTMTHINFYGVDVDENPELCE 86
FD A+KN L +VDF ATWCGPC+ I P+ L EK ++ F VDVD+ ++
Sbjct 11 FDNALKNAGDKLVVVDFTATWCGPCQTIGPYFKLLSEKPEN-KNVVFLKVDVDDAQDVAA 69
Query 87 REKVQAMPTFKLYKDGKVVETIVGVLKPKLEEALNNLK 124
+ MPTF YK+GK V+ G + KLEE +N+ K
Sbjct 70 LCGISCMPTFHFYKNGKKVDEFSGSNQSKLEEKINSHK 107
> ath:AT3G51030 ATTRX1; ATTRX1; oxidoreductase, acting on sulfur
group of donors, disulfide as acceptor
Length=114
Score = 74.3 bits (181), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 33/96 (34%), Positives = 56/96 (58%), Gaps = 5/96 (5%)
Query 30 IEEFDEAIKNG-----LAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDENPEL 84
+E ++E ++ L +VDF A+WCGPC+ I PF + + + ++ F VD DE +
Sbjct 14 VETWNEQLQKANESKTLVVVDFTASWCGPCRFIAPFFADLAKKLPNVLFLKVDTDELKSV 73
Query 85 CEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEAL 120
+QAMPTF K+GK+++ +VG K +L+ +
Sbjct 74 ASDWAIQAMPTFMFLKEGKILDKVVGAKKDELQSTI 109
> ath:AT1G45145 ATTRX5; ATTRX5; oxidoreductase, acting on sulfur
group of donors, disulfide as acceptor
Length=118
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 34/96 (35%), Positives = 58/96 (60%), Gaps = 5/96 (5%)
Query 30 IEEFDEAIKNG-----LAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDENPEL 84
+E ++E +K+ L ++DF A+WC PC+ I P E + T++ F+ +DVDE +
Sbjct 13 LEVWNEKVKDANESKKLIVIDFTASWCPPCRFIAPVFAEMAKKFTNVVFFKIDVDELQAV 72
Query 85 CEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEAL 120
+ KV+AMPTF K+G +++ +VG K ++ E L
Sbjct 73 AQEFKVEAMPTFVFMKEGNIIDRVVGAAKDEINEKL 108
> ath:AT5G42980 ATTRX3; ATTRX3 (THIOREDOXIN 3); oxidoreductase,
acting on sulfur group of donors, disulfide as acceptor
Length=118
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 40/100 (40%), Positives = 58/100 (58%), Gaps = 8/100 (8%)
Query 30 IEEFDEAIKNG-----LAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDENPEL 84
+E++ E +K L ++DF ATWC PC+ I P + + + F+ VDVDE +
Sbjct 13 VEDWTEKLKAANESKKLIVIDFTATWCPPCRFIAPVFADLAKKHLDVVFFKVDVDELNTV 72
Query 85 CEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEALNNLK 124
E KVQAMPTF K+G++ ET+VG K EE + NL+
Sbjct 73 AEEFKVQAMPTFIFMKEGEIKETVVGAAK---EEIIANLE 109
> ath:AT1G43560 Aty2; Aty2 (Arabidopsis thioredoxin y2); electron
carrier/ protein disulfide oxidoreductase
Length=167
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 37/100 (37%), Positives = 59/100 (59%), Gaps = 5/100 (5%)
Query 18 RLAKMPVKHLK--GIEEFDEAIKNG--LAIVDFFATWCGPCKMIRPFLEEKSNTMTH-IN 72
R A + V+ K FD+ ++N +VDF+ATWCGPC+++ P L E S T+ I
Sbjct 51 RFAPLTVRAAKKQTFNSFDDLLQNSDKPVLVDFYATWCGPCQLMVPILNEVSETLKDIIA 110
Query 73 FYGVDVDENPELCEREKVQAMPTFKLYKDGKVVETIVGVL 112
+D ++ P L + +++A+PTF L+KDGK+ + G L
Sbjct 111 VVKIDTEKYPSLANKYQIEALPTFILFKDGKLWDRFEGAL 150
> dre:336637 TXN, fb15h09, wu:fb15h09; zgc:56493; K03671 thioredoxin
1
Length=108
Score = 71.6 bits (174), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 38/99 (38%), Positives = 57/99 (57%), Gaps = 5/99 (5%)
Query 31 EEFDEAIKNG---LAIVDFFATWCGPCKMIRPFLEEKSNTMTHIN--FYGVDVDENPELC 85
+ FD+A+ L +VDF ATWCGPC+ I PF + S + N F VDVD+ ++
Sbjct 9 DGFDKALAGAGDKLVVVDFTATWCGPCQSIAPFYKGLSENPDYSNVVFLKVDVDDAQDVA 68
Query 86 EREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEALNNLK 124
+ +++ MPTF YK+GK ++ G + KLEE + K
Sbjct 69 QSCEIKCMPTFHFYKNGKKLDDFSGSNQTKLEEMVKQHK 107
> hsa:84203 TXNDC2, DKFZp434H0311, MGC35026, SPTRX, SPTRX1; thioredoxin
domain containing 2 (spermatozoa)
Length=553
Score = 70.9 bits (172), Expect = 1e-12, Method: Composition-based stats.
Identities = 40/104 (38%), Positives = 55/104 (52%), Gaps = 3/104 (2%)
Query 24 VKHLKGIEEFDEAIKNG---LAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDE 80
VK + E+F+ ++K L VDF ATWCGPC+ IRPF S + F VD D
Sbjct 450 VKVILSKEDFEASLKEAGERLVAVDFSATWCGPCRTIRPFFHALSVKHEDVVFLEVDADN 509
Query 81 NPELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEALNNLK 124
E+ + +PTF+ YK + V+ + G LK KLE + LK
Sbjct 510 CEEVVRECAIMCVPTFQFYKKEEKVDELCGALKEKLEAVIAELK 553
> cel:F56G4.5 png-1; PNG (Peptide:N-Glycanase) homolog family
member (png-1); K01456 peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine
amidase [EC:3.5.1.52]
Length=606
Score = 70.9 bits (172), Expect = 1e-12, Method: Composition-based stats.
Identities = 34/103 (33%), Positives = 55/103 (53%), Gaps = 4/103 (3%)
Query 22 MPVKHLKGIEEFDEAIK----NGLAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVD 77
MPV + + E + ++ N L I+DFFA WCGPC+MI P E+ S + F V+
Sbjct 1 MPVTEVGSLPELNNILERSDANRLIIIDFFANWCGPCRMISPIFEQFSAEYGNATFLKVN 60
Query 78 VDENPELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEAL 120
D ++ +R + AMPTF K+ + V+ + G + + E +
Sbjct 61 CDVARDIVQRYNISAMPTFIFLKNRQQVDMVRGANQQAIAEKI 103
> ath:AT3G08710 ATH9; ATH9 (thioredoxin H-type 9)
Length=140
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 33/94 (35%), Positives = 57/94 (60%), Gaps = 1/94 (1%)
Query 31 EEFDEAIKNG-LAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDENPELCEREK 89
++ EA ++G + + +F ATWCGPCK++ PF E S + + F VDVDE +
Sbjct 36 DKLAEADRDGKIVVANFSATWCGPCKIVAPFFIELSEKHSSLMFLLVDVDELSDFSSSWD 95
Query 90 VQAMPTFKLYKDGKVVETIVGVLKPKLEEALNNL 123
++A PTF K+G+ + +VG KP+L++ + ++
Sbjct 96 IKATPTFFFLKNGQQIGKLVGANKPELQKKVTSI 129
> mmu:213272 Txndc2, AU021712, Sptrx-1, Sptrx1, Trx4; thioredoxin
domain containing 2 (spermatozoa)
Length=550
Score = 68.9 bits (167), Expect = 3e-12, Method: Composition-based stats.
Identities = 37/104 (35%), Positives = 59/104 (56%), Gaps = 3/104 (2%)
Query 24 VKHLKGIEEFDEAIKNG---LAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDE 80
V+ +K EEF+E +K+ L VDF A WCGPC+M++P S + F VD ++
Sbjct 447 VRVIKDKEEFEEVLKDAGEKLVAVDFSAAWCGPCRMMKPLFHSLSLKHEDVIFLEVDTED 506
Query 81 NPELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEALNNLK 124
+L + ++ +PTF+ YK+ + V G L KLE +++ LK
Sbjct 507 CEQLVQDCEIFHLPTFQFYKNEEKVGEFSGALVGKLERSISELK 550
> eco:b3781 trxA, dasC, ECK3773, fipA, JW5856, tsnC; thioredoxin
1; K03671 thioredoxin 1
Length=109
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 35/95 (36%), Positives = 58/95 (61%), Gaps = 4/95 (4%)
Query 31 EEFDEAI--KNGLAIVDFFATWCGPCKMIRPFLEEKSNT-MTHINFYGVDVDENPELCER 87
+ FD + +G +VDF+A WCGPCKMI P L+E ++ + +++D+NP +
Sbjct 11 DSFDTDVLKADGAILVDFWAEWCGPCKMIAPILDEIADEYQGKLTVAKLNIDQNPGTAPK 70
Query 88 EKVQAMPTFKLYKDGKVVETIVGVL-KPKLEEALN 121
++ +PT L+K+G+V T VG L K +L+E L+
Sbjct 71 YGIRGIPTLLLFKNGEVAATKVGALSKGQLKEFLD 105
> bbo:BBOV_III011040 17.m07951; thioredoxin domain containing
protein
Length=128
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 34/94 (36%), Positives = 54/94 (57%), Gaps = 1/94 (1%)
Query 24 VKHLKGIEEFDEAIKNG-LAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDENP 82
VK K +E+ A+K + F A+WC PC+ + ++EE S + F VDVDE P
Sbjct 25 VKLAKSTDEYHAALKASEYVVAKFGASWCKPCQKAKVYVEELSEEHPELLFLDVDVDELP 84
Query 83 ELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKL 116
++ + E V ++P FK++K GK V+ +G K +L
Sbjct 85 QIADEEGVNSIPMFKMFKYGKAVDIHIGGDKQQL 118
> ath:AT1G76760 ATY1; ATY1 (ARABIDOPSIS THIOREDOXIN Y1); electron
carrier/ protein disulfide oxidoreductase
Length=172
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 29/74 (39%), Positives = 46/74 (62%), Gaps = 1/74 (1%)
Query 43 IVDFFATWCGPCKMIRPFLEEKSNTMT-HINFYGVDVDENPELCEREKVQAMPTFKLYKD 101
+VD++ATWCGPC+ + P L E S T+ I +D ++ P + + K++A+PTF L+KD
Sbjct 85 LVDYYATWCGPCQFMVPILNEVSETLKDKIQVVKIDTEKYPSIANKYKIEALPTFILFKD 144
Query 102 GKVVETIVGVLKPK 115
G+ + G L K
Sbjct 145 GEPCDRFEGALTAK 158
> cel:Y54E10A.3 hypothetical protein
Length=284
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 43/103 (41%), Positives = 54/103 (52%), Gaps = 11/103 (10%)
Query 22 MPVKHLKGIEEFDEAIK-NGL--AIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDV 78
MPV ++K E+F + GL IVDF A WCGPCKMI P E SN F VDV
Sbjct 1 MPVINVKDDEDFRNQLSLAGLKSVIVDFTAVWCGPCKMIAPTFEALSNQYLGAVFLKVDV 60
Query 79 DENPELCEREK----VQAMPTFKLYKDGKVVETIVGVLKPKLE 117
E+CE+ V +MPTF +++ G VE + G LE
Sbjct 61 ----EICEKTSSENGVNSMPTFMVFQSGVRVEQMKGADAKALE 99
> pfa:PFI1250w tlp2; Tlp2
Length=128
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 30/82 (36%), Positives = 49/82 (59%), Gaps = 0/82 (0%)
Query 39 NGLAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDENPELCEREKVQAMPTFKL 98
N L + F A+WC PCK ++P +E+ +I +D+DE PEL E E + +PT L
Sbjct 41 NKLVVAQFGASWCAPCKKMKPVIEKLGEDNDNIESLYIDIDEFPELGENEDINELPTILL 100
Query 99 YKDGKVVETIVGVLKPKLEEAL 120
K+GK ++ I+G+ + L +A+
Sbjct 101 RKNGKYLDKIIGMNESDLIKAV 122
> ath:AT1G19730 ATTRX4; ATTRX4; oxidoreductase, acting on sulfur
group of donors, disulfide as acceptor
Length=119
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 33/80 (41%), Positives = 48/80 (60%), Gaps = 1/80 (1%)
Query 39 NGLAIVDFFATWCGPCKMIRPFLEEKSNT-MTHINFYGVDVDENPELCEREKVQAMPTFK 97
N L ++DF A+WC PC+MI P + + M+ F+ VDVDE + + V+AMPTF
Sbjct 28 NKLIVIDFTASWCPPCRMIAPIFNDLAKKFMSSAIFFKVDVDELQSVAKEFGVEAMPTFV 87
Query 98 LYKDGKVVETIVGVLKPKLE 117
K G+VV+ +VG K L+
Sbjct 88 FIKAGEVVDKLVGANKEDLQ 107
> tpv:TP04_0217 thioredoxin; K03671 thioredoxin 1
Length=101
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 32/98 (32%), Positives = 55/98 (56%), Gaps = 1/98 (1%)
Query 24 VKHLKGIEEFDEAIK-NGLAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDENP 82
V + EEF++ + + + +VDF+A WCGPC P + + + F V+VD+
Sbjct 2 VHEVTSKEEFEKTLSGDSVVVVDFYADWCGPCMRFAPQFDALATEHPSLLFVKVNVDKLQ 61
Query 83 ELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEAL 120
EL ++ V ++PTFK++K G+V+ +G K L+ L
Sbjct 62 ELAQKYNVTSLPTFKVFKSGQVLGEFLGASKEGLKNTL 99
> mmu:67402 Txndc8, 4930429J24Rik, MGC151140, MGC151142, Sptrx-3,
Sptrx3, Trx6; thioredoxin domain containing 8
Length=153
Score = 65.1 bits (157), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 31/80 (38%), Positives = 44/80 (55%), Gaps = 3/80 (3%)
Query 24 VKHLKGIEEFDEAIK---NGLAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDE 80
VK +K + E E N L +V+F A WCGPCK I P + S ++ F VDVD
Sbjct 28 VKRIKNMSELKELFSDAGNKLVVVEFSAKWCGPCKTIAPVFQAMSLKYQNVTFAQVDVDS 87
Query 81 NPELCEREKVQAMPTFKLYK 100
+ EL E + +PTF+++K
Sbjct 88 SKELAEHCDITMLPTFQMFK 107
> cel:Y55F3AR.2 hypothetical protein
Length=254
Score = 64.7 bits (156), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 38/100 (38%), Positives = 50/100 (50%), Gaps = 3/100 (3%)
Query 22 MPVKHLKGIEEFDE--AIKNGLAI-VDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDV 78
MPV + G +FD + NG A+ VDF A+WCGPC+ I P + +N F VDV
Sbjct 1 MPVIVVNGDSDFDRKFSAGNGKAVFVDFTASWCGPCQYIAPIFSDLANQYKGSVFLKVDV 60
Query 79 DENPELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEE 118
DE V AMPTF + +G+ TI G + L
Sbjct 61 DECRGTAATYGVNAMPTFIAFVNGQKKATIQGADESGLRS 100
> tpv:TP01_0471 thioredoxin; K03671 thioredoxin 1
Length=248
Score = 64.3 bits (155), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 33/83 (39%), Positives = 50/83 (60%), Gaps = 8/83 (9%)
Query 38 KNGLAIVDFFATWCGPCKMIRPFLE-----EKSNTMTHINFYGVDVDENPELCEREKVQA 92
NG+ +VDFFATWCGPC+ + LE +S+ +T + VDVD+N E K+ A
Sbjct 154 SNGVLLVDFFATWCGPCQRMNENLEVIRSQYRSDKLT---IHSVDVDQNKSHLEEYKITA 210
Query 93 MPTFKLYKDGKVVETIVGVLKPK 115
+P+ L+ G+V + IVG++ K
Sbjct 211 LPSLLLFVRGEVHKKIVGLVDVK 233
> ath:AT3G53220 thioredoxin family protein
Length=126
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 26/79 (32%), Positives = 46/79 (58%), Gaps = 2/79 (2%)
Query 42 AIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDENPELCEREKVQAMPTFKLYKD 101
A++++ A+WCG C I P + SN+ + + F D+DE PE ++ PTF+ Y+D
Sbjct 46 AVINYGASWCGVCSQILPAFRKLSNSFSKLKFVYADIDECPETTRH--IRYTPTFQFYRD 103
Query 102 GKVVETIVGVLKPKLEEAL 120
G+ V+ + G + +L + L
Sbjct 104 GEKVDEMFGAGEQRLHDRL 122
> ath:AT4G26160 ACHT1; ACHT1 (ATYPICAL CYS HIS RICH THIOREDOXIN
1); oxidoreductase, acting on sulfur group of donors, disulfide
as acceptor
Length=221
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 36/100 (36%), Positives = 54/100 (54%), Gaps = 6/100 (6%)
Query 27 LKGIEEFDEAIKNG---LAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDENPE 83
+ E+F A+K+ L IVDF+ TWCG C+ + P L + + +I F V+ DEN
Sbjct 98 ITSAEQFLNALKDAGDRLVIVDFYGTWCGSCRAMFPKLCKTAKEHPNILFLKVNFDENKS 157
Query 84 LCEREKVQAMPTFKLYK--DGKVVETIVGVLK-PKLEEAL 120
LC+ V+ +P F Y+ DG+V + K KL EA+
Sbjct 158 LCKSLNVKVLPYFHFYRGADGQVESFSCSLAKFQKLREAI 197
> ath:AT1G03680 ATHM1; ATHM1; enzyme activator
Length=179
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 48/80 (60%), Gaps = 2/80 (2%)
Query 44 VDFFATWCGPCKMIRPFLEEKSNTMT-HINFYGVDVDENPELCEREKVQAMPTFKLYKDG 102
VDF+A WCGPCKMI P + E + FY ++ DE+P + V+++PT ++ +G
Sbjct 97 VDFWAPWCGPCKMIDPIVNELAQKYAGQFKFYKLNTDESPATPGQYGVRSIPTIMIFVNG 156
Query 103 KVVETIVG-VLKPKLEEALN 121
+ +TI+G V K L ++N
Sbjct 157 EKKDTIIGAVSKDTLATSIN 176
> ath:AT3G17880 ATTDX; ATTDX (TETRATICOPEPTIDE DOMAIN-CONTAINING
THIOREDOXIN); oxidoreductase, acting on sulfur group of donors,
disulfide as acceptor / protein binding
Length=373
Score = 61.2 bits (147), Expect = 9e-10, Method: Composition-based stats.
Identities = 27/80 (33%), Positives = 45/80 (56%), Gaps = 0/80 (0%)
Query 41 LAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDENPELCEREKVQAMPTFKLYK 100
L I+ F ATWCGPC+ + P + + + F VD+D+ ++ + ++PTF +
Sbjct 287 LLILYFTATWCGPCRYMSPLYSNLATQHSRVVFLKVDIDKANDVAASWNISSVPTFCFIR 346
Query 101 DGKVVETIVGVLKPKLEEAL 120
DGK V+ +VG K LE+ +
Sbjct 347 DGKEVDKVVGADKGSLEQKI 366
> ath:AT3G15360 TRX-M4; TRX-M4 (ARABIDOPSIS THIOREDOXIN M-TYPE
4); enzyme activator
Length=193
Score = 61.2 bits (147), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 45/71 (63%), Gaps = 1/71 (1%)
Query 43 IVDFFATWCGPCKMIRPFLEEKSNTMT-HINFYGVDVDENPELCEREKVQAMPTFKLYKD 101
+V+F+A WCGPC+MI P +++ + FY ++ DE+P R ++++PT ++K
Sbjct 108 LVEFWAPWCGPCRMIHPIVDQLAKDFAGKFKFYKINTDESPNTANRYGIRSVPTVIIFKG 167
Query 102 GKVVETIVGVL 112
G+ ++I+G +
Sbjct 168 GEKKDSIIGAV 178
> ath:AT1G50320 ATHX; ATHX; enzyme activator
Length=182
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 34/85 (40%), Positives = 47/85 (55%), Gaps = 4/85 (4%)
Query 43 IVDFFATWCGPCKMIRPFLEEKSNTMT-HINFYGVDVDENPELCEREKVQAMPTFKLYKD 101
+V+F ATWCGPCK+I P +E S + +D D NP+L KV +P F L+KD
Sbjct 91 LVEFVATWCGPCKLIYPAMEALSQEYGDKLTIVKIDHDANPKLIAEFKVYGLPHFILFKD 150
Query 102 GKVVETIV---GVLKPKLEEALNNL 123
GK V + K KL+E ++ L
Sbjct 151 GKEVPGSRREGAITKAKLKEYIDGL 175
> ath:AT1G69880 ATH8; ATH8 (thioredoxin H-type 8)
Length=148
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 56/113 (49%), Gaps = 5/113 (4%)
Query 14 IFDFRLAKMPVKHLKGIEEFDEAIK-----NGLAIVDFFATWCGPCKMIRPFLEEKSNTM 68
I+ F++ + +K + ++ + N L +++F A WCGPCK + P LEE +
Sbjct 29 IYPFKVNSPCIVEIKNMNQWKSRLNALKDTNKLLVIEFTAKWCGPCKTLEPKLEELAAKY 88
Query 69 THINFYGVDVDENPELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEALN 121
T + F +DVD + + +P K G+ V+ +VGV +LE LN
Sbjct 89 TDVEFVKIDVDVLMSVWMEFNLSTLPAIVFMKRGREVDMVVGVKVDELERKLN 141
> bbo:BBOV_I003690 19.m02235; thioredoxin
Length=253
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 31/96 (32%), Positives = 52/96 (54%), Gaps = 7/96 (7%)
Query 34 DEAIKNGLAIVDFFATWCGPCKMIRPFLE--EKSNTMTHINFYGVDVDENPELCEREKVQ 91
D G+ +VDF+ATWC PC ++ L E + + +DVD+N EL ++
Sbjct 156 DNKTSGGVLLVDFYATWCRPCDTMKANLSIIESRYKPGKLVIHKIDVDQNAELSASYEIT 215
Query 92 AMPTFKLYKDGKVVETIVGV-----LKPKLEEALNN 122
A+PT L++ G++V+ + GV L + EAL++
Sbjct 216 ALPTLLLFRRGELVKRVRGVVDVSTLMGMINEALDD 251
> ath:AT1G31020 ATO2; ATO2 (Arabidopsis thioredoxin o2)
Length=159
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 34/104 (32%), Positives = 50/104 (48%), Gaps = 7/104 (6%)
Query 27 LKGIEEFDEAIKNGL-----AIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDEN 81
LK EF+ A+ ++ F A WCGPC++I P + E SN + Y VD+DE
Sbjct 54 LKSEAEFNSALSKARDGSLPSVFYFTAAWCGPCRLISPVILELSNKYPDVTTYKVDIDEG 113
Query 82 --PELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEALNNL 123
+ V A+PT + +K G IVGV +L+ + L
Sbjct 114 GLSNAIGKLNVSAVPTLQFFKGGVKKAEIVGVDVVRLKSVMEQL 157
> eco:b2582 trxC, ECK2580, JW2566, yfiG; thioredoxin 2 (EC:1.8.1.8);
K03672 thioredoxin 2 [EC:1.8.1.8]
Length=139
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 31/94 (32%), Positives = 54/94 (57%), Gaps = 3/94 (3%)
Query 31 EEFDEAIKNGLAIV-DFFATWCGPCKMIRPFLEEKSNTMT-HINFYGVDVDENPELCERE 88
E D+ +K+ L +V DF+A WCGPC+ P E+ + + + F V+ + EL R
Sbjct 43 ETLDKLLKDDLPVVIDFWAPWCGPCRNFAPIFEDVAQERSGKVRFVKVNTEAERELSSRF 102
Query 89 KVQAMPTFKLYKDGKVVETIVG-VLKPKLEEALN 121
++++PT ++K+G+VV+ + G V K + LN
Sbjct 103 GIRSIPTIMIFKNGQVVDMLNGAVPKAPFDSWLN 136
> dre:402938 txn2, MGC77127, zgc:77127; thioredoxin 2; K03671
thioredoxin 1
Length=166
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 46/84 (54%), Gaps = 3/84 (3%)
Query 31 EEFDEAIKNG--LAIVDFFATWCGPCKMIRPFLEEK-SNTMTHINFYGVDVDENPELCER 87
++F E + N ++DF A WCGPCK++ P LE+ + + VD+DE+ +L
Sbjct 67 DDFTERVINSELPVLIDFHAQWCGPCKILGPRLEKAIAKQKGRVTMAKVDIDEHTDLAIE 126
Query 88 EKVQAMPTFKLYKDGKVVETIVGV 111
V A+PT + G V++ VG+
Sbjct 127 YGVSAVPTVIAMRGGDVIDQFVGI 150
> ath:AT2G40790 ATCXXS2 (C-TERMINAL CYSTEINE RESIDUE IS CHANGED
TO A SERINE 1); protein disulfide isomerase
Length=154
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 33/90 (36%), Positives = 50/90 (55%), Gaps = 2/90 (2%)
Query 23 PVKHLKGIEE-FDEAIKNG-LAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDE 80
PV ++ EE EA +G + +V+F A+WC P K I P +E ++T T + F +DV+E
Sbjct 44 PVSRMEKWEEKITEANSHGKILVVNFKASWCLPSKTILPIYQELASTYTSMIFVTIDVEE 103
Query 81 NPELCEREKVQAMPTFKLYKDGKVVETIVG 110
E V A PT KDG+ ++ +VG
Sbjct 104 LAEFSHEWNVDATPTVVFLKDGRQMDKLVG 133
> ath:AT1G53300 TTL1; TTL1 (TETRATRICOPETIDE-REPEAT THIOREDOXIN-LIKE
1); binding
Length=699
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 31/100 (31%), Positives = 54/100 (54%), Gaps = 1/100 (1%)
Query 24 VKHLKGIEEFDEAIK-NGLAIVDFFATWCGPCKMIRPFLEEKSNTMTHINFYGVDVDENP 82
V+ + +E+F A+ G++++ F CK I PF++ I+F VD+D+ P
Sbjct 597 VEEIYSLEQFKSAMNLPGVSVIHFSTASDHQCKQISPFVDSLCTRYPSIHFLKVDIDKCP 656
Query 83 ELCEREKVQAMPTFKLYKDGKVVETIVGVLKPKLEEALNN 122
+ E V+ +PT K+YK+G V+ IV K LE ++ +
Sbjct 657 SIGNAENVRVVPTVKIYKNGSRVKEIVCPSKEVLEYSVRH 696
> cel:B0024.9 trx-2; ThioRedoXin [see also xtr] family member
(trx-2); K03671 thioredoxin 1
Length=145
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/89 (34%), Positives = 46/89 (51%), Gaps = 3/89 (3%)
Query 27 LKGIEEFDEAIKNGLA--IVDFFATWCGPCKMIRPFLEEKSNT-MTHINFYGVDVDENPE 83
+ +E+F E + IVDF A WCGPC+ + P LEEK N + ++VD E
Sbjct 42 IDSVEDFTEKVIQSSVPVIVDFHAEWCGPCQALGPRLEEKVNGRQGSVLLAKINVDHAGE 101
Query 84 LCEREKVQAMPTFKLYKDGKVVETIVGVL 112
L + A+PT +K+G+ + GVL
Sbjct 102 LAMDYGISAVPTVFAFKNGEKISGFSGVL 130
Lambda K H
0.323 0.141 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2069971060
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40