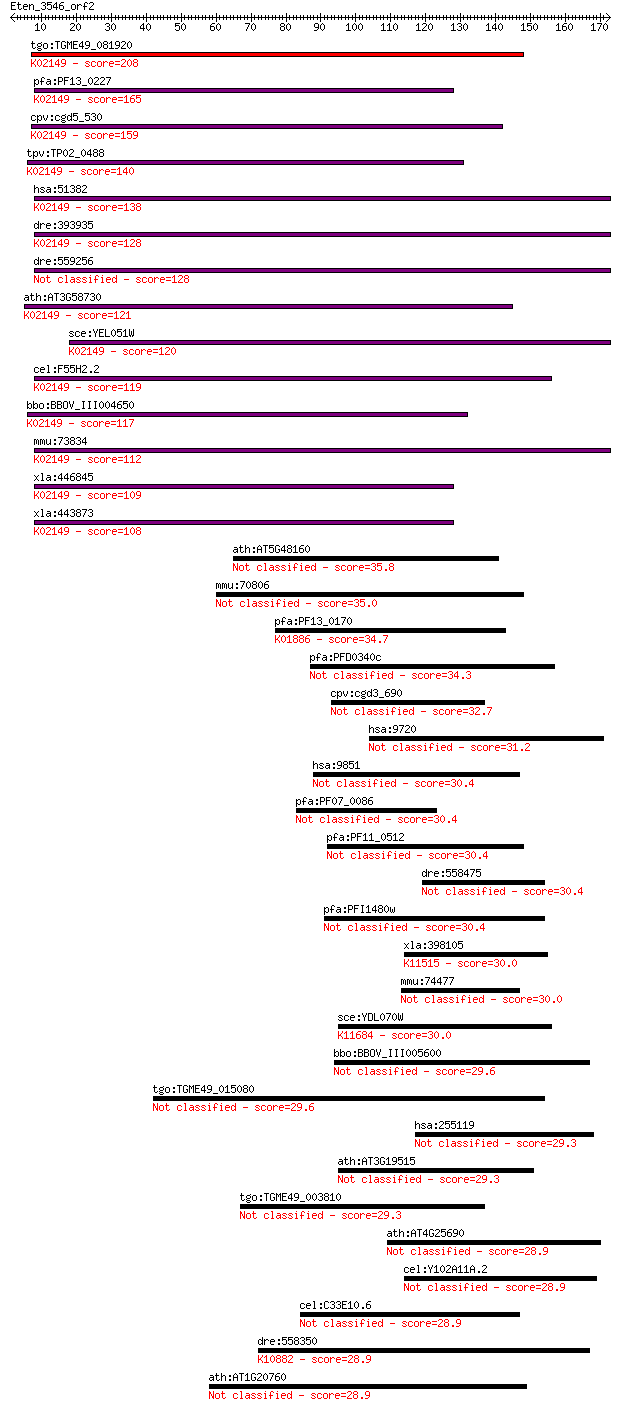
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3546_orf2
Length=172
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_081920 vacuolar ATP synthase subunit D, putative (E... 208 9e-54
pfa:PF13_0227 vacuolar ATP synthase subunit D, putative (EC:3.... 165 8e-41
cpv:cgd5_530 vacuolar H-ATpase subunit D ; K02149 V-type H+-tr... 159 3e-39
tpv:TP02_0488 vacuolar ATP synthase subunit D; K02149 V-type H... 140 3e-33
hsa:51382 ATP6V1D, ATP6M, VATD, VMA8; ATPase, H+ transporting,... 138 7e-33
dre:393935 atp6v1d, MGC55524, zgc:55524; ATPase, H+ transporti... 128 1e-29
dre:559256 atp6v1d; ATPase, H+ transporting, V1 subunit D 128 1e-29
ath:AT3G58730 vacuolar ATP synthase subunit D (VATD) / V-ATPas... 121 1e-27
sce:YEL051W VMA8; Subunit D of the eight-subunit V1 peripheral... 120 2e-27
cel:F55H2.2 vha-14; Vacuolar H ATPase family member (vha-14); ... 119 5e-27
bbo:BBOV_III004650 17.m07416; V-type ATPase, D subunit family ... 117 3e-26
mmu:73834 Atp6v1d, 1110004P10Rik, Atp6m, VATD, Vma8; ATPase, H... 112 4e-25
xla:446845 atp6v1d, MGC80692; ATPase, H+ transporting, lysosom... 109 4e-24
xla:443873 MGC79146 protein; K02149 V-type H+-transporting ATP... 108 1e-23
ath:AT5G48160 OBE2; OBE2 (OBERON2); protein binding / zinc ion... 35.8 0.071
mmu:70806 Wdr96, 4632415N18Rik, 4930428C11Rik, 4930463G05Rik, ... 35.0 0.13
pfa:PF13_0170 glutaminyl-tRNA synthetase, putative (EC:6.1.1.1... 34.7 0.17
pfa:PFD0340c conserved Plasmodium protein, unknown function 34.3 0.19
cpv:cgd3_690 ABC transporter 32.7 0.64
hsa:9720 CCDC144A, FLJ43983, KIAA0565, MGC164650; coiled-coil ... 31.2 1.6
hsa:9851 MGC130040, MGC130041; KIAA0753 30.4 2.7
pfa:PF07_0086 conserved Plasmodium membrane protein, unknown f... 30.4 2.8
pfa:PF11_0512 RESA-like protein with PHIST and DnaJ domains 30.4 2.8
dre:558475 si:ch211-57h10.1 30.4 3.1
pfa:PFI1480w conserved Plasmodium protein, unknown function 30.4 3.4
xla:398105 incenp-a, xl-incenp; inner centromere protein antig... 30.0 3.8
mmu:74477 4933427D14Rik, C85113, Kiaa0753; RIKEN cDNA 4933427D... 30.0 4.2
sce:YDL070W BDF2; Bdf2p; K11684 bromodomain-containing factor 1 30.0
bbo:BBOV_III005600 17.m07500; Spherical Body Protein 2 truncat... 29.6 4.9
tgo:TGME49_015080 hypothetical protein 29.6 5.1
hsa:255119 C4orf22, MGC35043; chromosome 4 open reading frame 22 29.3
ath:AT3G19515 hypothetical protein 29.3 7.2
tgo:TGME49_003810 kelch motif domain-containing protein 29.3 7.4
ath:AT4G25690 hypothetical protein 28.9 8.2
cel:Y102A11A.2 hypothetical protein 28.9 8.3
cel:C33E10.6 hypothetical protein 28.9 8.5
dre:558350 eme1, fc30c07, wu:fc30c07; essential meiotic endonu... 28.9 8.8
ath:AT1G20760 calcium-binding EF hand family protein 28.9 9.4
> tgo:TGME49_081920 vacuolar ATP synthase subunit D, putative
(EC:3.6.3.14); K02149 V-type H+-transporting ATPase subunit
D [EC:3.6.3.14]
Length=245
Score = 208 bits (529), Expect = 9e-54, Method: Compositional matrix adjust.
Identities = 97/141 (68%), Positives = 118/141 (83%), Gaps = 0/141 (0%)
Query 7 FLQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEA 66
L+R++RPA F+ A DNVAGV LP+F I TDP+VD+LKN+ ++AGG VI+AAR+ + +
Sbjct 81 LLERVRRPATFLNVAADNVAGVTLPIFHICTDPSVDVLKNVGVAAGGQVIMAAREMFLKV 140
Query 67 LAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFR 126
+ELVKLASLQTAFFTLD EIKMTNRRVNAL+NVVLP++D INYI +ELDEMEREEFFR
Sbjct 141 FSELVKLASLQTAFFTLDEEIKMTNRRVNALSNVVLPRIDGGINYIVRELDEMEREEFFR 200
Query 127 LKKIQEKKRLAKEAEQKALEE 147
LKKIQEKKR+ KE E + L E
Sbjct 201 LKKIQEKKRMRKEEEDRLLHE 221
> pfa:PF13_0227 vacuolar ATP synthase subunit D, putative (EC:3.6.3.14);
K02149 V-type H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=247
Score = 165 bits (417), Expect = 8e-41, Method: Compositional matrix adjust.
Identities = 76/120 (63%), Positives = 97/120 (80%), Gaps = 0/120 (0%)
Query 8 LQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEAL 67
++ IKRP V ++ + +NVAGV+LP+FQ+ DPTVD+L N+ ++AGG VI R+ Y + L
Sbjct 84 IEGIKRPVVTLSLSTNNVAGVKLPIFQVNIDPTVDVLGNLGVAAGGQVINNTRENYLQCL 143
Query 68 AELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRL 127
LVKLAS+Q AFF+LD EIKMTNRRVNALNN+VLP+LD INYI KELDE+EREEF+RL
Sbjct 144 NMLVKLASMQVAFFSLDEEIKMTNRRVNALNNIVLPRLDGGINYIIKELDEIEREEFYRL 203
> cpv:cgd5_530 vacuolar H-ATpase subunit D ; K02149 V-type H+-transporting
ATPase subunit D [EC:3.6.3.14]
Length=249
Score = 159 bits (403), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 79/136 (58%), Positives = 101/136 (74%), Gaps = 1/136 (0%)
Query 7 FLQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDI-LKNINLSAGGHVILAARDKYQE 65
++ KRP V + +N+AGVRLP+F++ D +I +++GG VI + R+ Y +
Sbjct 74 IIESCKRPTVTMEVGTENIAGVRLPIFEMNVDNNSSTETCHIGVASGGQVIQSTREIYMK 133
Query 66 ALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFF 125
L +LVKLASLQTAFF+LD EIKMTNRRVNAL NVVLPKL+ +NYI +ELDE+EREEFF
Sbjct 134 VLRDLVKLASLQTAFFSLDEEIKMTNRRVNALQNVVLPKLEDGMNYILRELDEIEREEFF 193
Query 126 RLKKIQEKKRLAKEAE 141
RLKKIQEKK+ EAE
Sbjct 194 RLKKIQEKKKEWAEAE 209
> tpv:TP02_0488 vacuolar ATP synthase subunit D; K02149 V-type
H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=238
Score = 140 bits (352), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 65/125 (52%), Positives = 94/125 (75%), Gaps = 0/125 (0%)
Query 6 LFLQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQE 65
L ++ + RP+V + +N+AGV LPVF + TDPTVD+ N++LS+GG I + + +
Sbjct 83 LVIESVGRPSVTLKLRGENIAGVLLPVFSLQTDPTVDLFANLSLSSGGSAIQSVKTTHLA 142
Query 66 ALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFF 125
AL LV+LASLQ +F L+ EI+MTNRR+NAL+NV++P +D+++ YI +ELDEMEREEF+
Sbjct 143 ALDILVELASLQISFIILNEEIRMTNRRINALDNVLIPSIDRNLEYIRRELDEMEREEFY 202
Query 126 RLKKI 130
RLK I
Sbjct 203 RLKMI 207
> hsa:51382 ATP6V1D, ATP6M, VATD, VMA8; ATPase, H+ transporting,
lysosomal 34kDa, V1 subunit D (EC:3.6.3.14 3.6.1.34); K02149
V-type H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=247
Score = 138 bits (348), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 78/168 (46%), Positives = 114/168 (67%), Gaps = 8/168 (4%)
Query 8 LQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEAL 67
+Q + + V + A DNVAGV LPVF+ + T D + L+ GG + + Y +A+
Sbjct 84 IQNVNKAQVKIRAKKDNVAGVTLPVFEHYHEGT-DSYELTGLARGGEQLAKLKRNYAKAV 142
Query 68 AELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRL 127
LV+LASLQT+F TLD IK+TNRRVNA+ +V++P++++++ YI ELDE EREEF+RL
Sbjct 143 ELLVELASLQTSFVTLDEAIKITNRRVNAIEHVIIPRIERTLAYIITELDEREREEFYRL 202
Query 128 KKIQEKKRLAKEAEQKALEE---AGKALKASGLLSGTILGEDDDDLVF 172
KKIQEKK++ KE +K LE+ AG+ L+ + LL+ E D+DL+F
Sbjct 203 KKIQEKKKILKEKSEKDLEQRRAAGEVLEPANLLA----EEKDEDLLF 246
> dre:393935 atp6v1d, MGC55524, zgc:55524; ATPase, H+ transporting,
V1 subunit D; K02149 V-type H+-transporting ATPase subunit
D [EC:3.6.3.14]
Length=248
Score = 128 bits (321), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 72/165 (43%), Positives = 106/165 (64%), Gaps = 1/165 (0%)
Query 8 LQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEAL 67
+Q + + V V A DNVAGV LPVF+ + D + L+ GG + + Y +A+
Sbjct 84 IQNVNKAQVKVRAKKDNVAGVTLPVFEHYQEGG-DSYELTGLARGGEQLSRLKRNYAKAV 142
Query 68 AELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRL 127
LV+LASLQT+F TLD IK+TNRRVNA+ +V++P++++++ YI ELDE EREEF+RL
Sbjct 143 ELLVELASLQTSFVTLDEAIKITNRRVNAIEHVIIPRIERTLTYIITELDEREREEFYRL 202
Query 128 KKIQEKKRLAKEAEQKALEEAGKALKASGLLSGTILGEDDDDLVF 172
KKIQEKK+ +E +K + + AL + + E D+DL+F
Sbjct 203 KKIQEKKKQLRERTEKEIAKRLAALGPIAEPTNMLTEEADEDLLF 247
> dre:559256 atp6v1d; ATPase, H+ transporting, V1 subunit D
Length=248
Score = 128 bits (321), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 72/165 (43%), Positives = 106/165 (64%), Gaps = 1/165 (0%)
Query 8 LQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEAL 67
+Q + + V V A DNVAGV LPVF+ + D + L+ GG + + Y +A+
Sbjct 84 IQNVNKAQVKVRAKKDNVAGVTLPVFEHYQEGG-DSYELTGLARGGEQLSRLKRNYAKAV 142
Query 68 AELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRL 127
LV+LASLQT+F TLD IK+TNRRVNA+ +V++P++++++ YI ELDE EREEF+RL
Sbjct 143 ELLVELASLQTSFVTLDEAIKITNRRVNAIEHVIIPRIERTLTYIITELDEREREEFYRL 202
Query 128 KKIQEKKRLAKEAEQKALEEAGKALKASGLLSGTILGEDDDDLVF 172
KKIQEKK+ +E +K + + AL + + E D+DL+F
Sbjct 203 KKIQEKKKQLRERTEKEIAKRLAALGPIAEPTNMLTEEADEDLLF 247
> ath:AT3G58730 vacuolar ATP synthase subunit D (VATD) / V-ATPase
D subunit / vacuolar proton pump D subunit (VATPD); K02149
V-type H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=261
Score = 121 bits (303), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 70/141 (49%), Positives = 94/141 (66%), Gaps = 5/141 (3%)
Query 5 WLFLQRIKRPAVFVTAAYDNVAGVRLPVF-QITTDPTVDILKNINLSAGGHVILAARDKY 63
+ L+ +K + V + +N+AGV+LP F + T + L L+ GG + A R Y
Sbjct 83 HVVLENVKEATLKVRSRTENIAGVKLPKFDHFSEGETKNDL--TGLARGGQQVRACRVAY 140
Query 64 QEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREE 123
+A+ LV+LASLQT+F TLD IK TNRRVNAL NVV PKL+ +I+YI ELDE+ERE+
Sbjct 141 VKAIEVLVELASLQTSFLTLDEAIKTTNRRVNALENVVKPKLENTISYIKGELDELERED 200
Query 124 FFRLKKIQEKKRLAKEAEQKA 144
FFRLKKIQ KR +E E++A
Sbjct 201 FFRLKKIQGYKR--REVERQA 219
> sce:YEL051W VMA8; Subunit D of the eight-subunit V1 peripheral
membrane domain of the vacuolar H+-ATPase (V-ATPase), an
electrogenic proton pump found throughout the endomembrane system;
plays a role in the coupling of proton transport and
ATP hydrolysis (EC:3.6.3.14); K02149 V-type H+-transporting
ATPase subunit D [EC:3.6.3.14]
Length=256
Score = 120 bits (302), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 69/163 (42%), Positives = 103/163 (63%), Gaps = 8/163 (4%)
Query 18 VTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAELVKLASLQ 77
V A +NV+GV L F+ DP ++ + L GG + A++ Y A+ LV+LASLQ
Sbjct 94 VRARQENVSGVYLSQFESYIDPEINDFRLTGLGRGGQQVQRAKEIYSRAVETLVELASLQ 153
Query 78 TAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQEKKR-- 135
TAF LD IK+TNRRVNA+ +V++P+ + +I YI ELDE++REEF+RLKK+QEKK+
Sbjct 154 TAFIILDEVIKVTNRRVNAIEHVIIPRTENTIAYINSELDELDREEFYRLKKVQEKKQNE 213
Query 136 LAK-EAEQK-----ALEEAGKALKASGLLSGTILGEDDDDLVF 172
AK +AE K A ++A + T++ + +DD++F
Sbjct 214 TAKLDAEMKLKRDRAEQDASEVAADEEPQGETLVADQEDDVIF 256
> cel:F55H2.2 vha-14; Vacuolar H ATPase family member (vha-14);
K02149 V-type H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=257
Score = 119 bits (298), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 69/150 (46%), Positives = 91/150 (60%), Gaps = 3/150 (2%)
Query 8 LQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEAL 67
+Q + + V +NV GV LPVF D D L GG I + Y +A+
Sbjct 86 IQNVSQAQYRVRMKKENVVGVFLPVFDAYQDGP-DAYDLTGLGKGGANIARLKKNYNKAI 144
Query 68 AELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRL 127
LV+LA+LQT F TLD IK+TNRRVNA+ +V++P+++ ++ YI ELDEMEREEFFR+
Sbjct 145 ELLVELATLQTCFITLDEAIKVTNRRVNAIEHVIIPRIENTLTYIVTELDEMEREEFFRM 204
Query 128 KKIQEKKRLAKEAE--QKALEEAGKALKAS 155
KKIQ K+ KE E QKALE G A+
Sbjct 205 KKIQANKKKLKEQEAAQKALEGPGPGEDAA 234
> bbo:BBOV_III004650 17.m07416; V-type ATPase, D subunit family
protein; K02149 V-type H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=233
Score = 117 bits (292), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 64/126 (50%), Positives = 92/126 (73%), Gaps = 0/126 (0%)
Query 6 LFLQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQE 65
L ++ + R AV + +NVAGV +P F++ DPTVD++ NI L+ GGHVI + + + E
Sbjct 68 LVVESVGRSAVTLRVRTENVAGVIIPHFELKIDPTVDVIANIGLTTGGHVIHSVKTAHLE 127
Query 66 ALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFF 125
L L +LASLQ +F L+ EIKMTNRRVNAL+N+V+P +D ++ YI +ELDE+EREEF+
Sbjct 128 FLETLAELASLQVSFMMLEQEIKMTNRRVNALDNLVIPTIDNNLEYIKRELDELEREEFY 187
Query 126 RLKKIQ 131
RLK ++
Sbjct 188 RLKMVR 193
> mmu:73834 Atp6v1d, 1110004P10Rik, Atp6m, VATD, Vma8; ATPase,
H+ transporting, lysosomal V1 subunit D; K02149 V-type H+-transporting
ATPase subunit D [EC:3.6.3.14]
Length=247
Score = 112 bits (281), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 77/168 (45%), Positives = 113/168 (67%), Gaps = 8/168 (4%)
Query 8 LQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEAL 67
+Q + + V + A DNVAGV LPVF+ + T D + L+ GG + + Y +A+
Sbjct 84 IQNVNKAQVKIRAKKDNVAGVTLPVFEHYHEGT-DSYELTGLARGGEQLAKLKRNYAKAV 142
Query 68 AELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRL 127
LV+LASLQT+F TLD IK+TNRRVNA+ +V++P++++++ YI ELDE EREEF+RL
Sbjct 143 ELLVELASLQTSFVTLDEAIKITNRRVNAIEHVIIPRIERTLAYIITELDEREREEFYRL 202
Query 128 KKIQEKKRLAKEAEQKALEE---AGKALKASGLLSGTILGEDDDDLVF 172
KKIQEKK++ KE +K LE AG+ ++ + LL+ E D+DL+F
Sbjct 203 KKIQEKKKIIKEKFEKDLERRRAAGEVMEPANLLA----EEKDEDLLF 246
> xla:446845 atp6v1d, MGC80692; ATPase, H+ transporting, lysosomal
34kDa, V1 subunit D (EC:3.6.3.14); K02149 V-type H+-transporting
ATPase subunit D [EC:3.6.3.14]
Length=248
Score = 109 bits (273), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 57/120 (47%), Positives = 82/120 (68%), Gaps = 1/120 (0%)
Query 8 LQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEAL 67
+Q + + V V A DNVAGV LPVF+ + D + L+ GG + + Y +A+
Sbjct 84 IQNVNKSQVKVRAKKDNVAGVTLPVFEHYQEGG-DSYELTGLARGGEQLAKLKRNYAKAV 142
Query 68 AELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRL 127
LV+LASLQT+F TLD IK+TNRRVNA+ +V++PK++++++YI ELDE EREEF+RL
Sbjct 143 ELLVELASLQTSFVTLDEAIKITNRRVNAIEHVIIPKIERTLSYIITELDEREREEFYRL 202
> xla:443873 MGC79146 protein; K02149 V-type H+-transporting ATPase
subunit D [EC:3.6.3.14]
Length=246
Score = 108 bits (269), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 56/120 (46%), Positives = 82/120 (68%), Gaps = 1/120 (0%)
Query 8 LQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEAL 67
+Q + + V V A DNVAGV LPVF+ + D + L+ GG + + Y +A+
Sbjct 84 IQNVNKAQVKVRARKDNVAGVTLPVFEHYQEGG-DSYELTGLARGGEQLAKLKRNYAKAV 142
Query 68 AELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRL 127
LV+LASLQT+F TLD IK+TNRRVNA+ +V++P+++++++YI ELDE EREEF+RL
Sbjct 143 ELLVELASLQTSFVTLDEAIKITNRRVNAIEHVIIPRIERTLSYIVTELDEREREEFYRL 202
> ath:AT5G48160 OBE2; OBE2 (OBERON2); protein binding / zinc ion
binding
Length=574
Score = 35.8 bits (81), Expect = 0.071, Method: Composition-based stats.
Identities = 25/83 (30%), Positives = 41/83 (49%), Gaps = 9/83 (10%)
Query 65 EALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKS-----INYITKELDEM 119
+ L +V+L + F L + R + L +VL K+DKS NY+ + L E
Sbjct 451 DELERIVRLKQAEADMFQLKA--NEAKREADRLQRIVLAKMDKSEEEYASNYLKQRLSEA 508
Query 120 EREEFFRLKKI--QEKKRLAKEA 140
E E+ + +KI QE R+A ++
Sbjct 509 EAEKQYLFEKIKLQENSRVASQS 531
> mmu:70806 Wdr96, 4632415N18Rik, 4930428C11Rik, 4930463G05Rik,
AI429486, D19Ertd652e; WD repeat domain 96
Length=1682
Score = 35.0 bits (79), Expect = 0.13, Method: Composition-based stats.
Identities = 39/97 (40%), Positives = 54/97 (55%), Gaps = 11/97 (11%)
Query 60 RDKYQEAL-AELVKLA-SLQTAFFTLDSEIK-MTNRRVNALNNVVLPKLDKSINYI--TK 114
RDKY+++L AEL KL S+Q + D +K + RRV A +V+ + + IN I +
Sbjct 1192 RDKYRKSLEAELKKLQNSIQESTQNFDDHLKRLFERRVKA--EMVINQEELKINNIIFSL 1249
Query 115 ELDE--MEREEFFR--LKKIQEKKRLAKEAEQKALEE 147
LDE RE+F L K QE+K EA QKA E+
Sbjct 1250 LLDEELSSREQFLNNYLLKKQEEKTKTAEAIQKARED 1286
> pfa:PF13_0170 glutaminyl-tRNA synthetase, putative (EC:6.1.1.18);
K01886 glutaminyl-tRNA synthetase [EC:6.1.1.18]
Length=918
Score = 34.7 bits (78), Expect = 0.17, Method: Composition-based stats.
Identities = 26/73 (35%), Positives = 42/73 (57%), Gaps = 9/73 (12%)
Query 77 QTAFFTLD----SEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFF---RLKK 129
+ FFT D +E+ + N V ++N +L K K + + KELD+++RE+ +LKK
Sbjct 847 RVGFFTKDKDTTNELPVFNLTVPLVDNTMLKK--KKEDLLQKELDKLKREKIAAERKLKK 904
Query 130 IQEKKRLAKEAEQ 142
Q+K R K+ EQ
Sbjct 905 EQKKIREQKKKEQ 917
> pfa:PFD0340c conserved Plasmodium protein, unknown function
Length=1751
Score = 34.3 bits (77), Expect = 0.19, Method: Composition-based stats.
Identities = 22/70 (31%), Positives = 37/70 (52%), Gaps = 7/70 (10%)
Query 87 IKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQEKKRLAKEAEQKALE 146
+K T R+VN NN +L + + I K+L ME + ++K ++KK + K E+
Sbjct 1523 VKRTKRKVNLSNNFIL----NNFSNILKKLQRMEED---KIKMDEQKKEINKNNEKGEFN 1575
Query 147 EAGKALKASG 156
E G+ +K G
Sbjct 1576 EKGEDIKEKG 1585
> cpv:cgd3_690 ABC transporter
Length=1586
Score = 32.7 bits (73), Expect = 0.64, Method: Composition-based stats.
Identities = 18/44 (40%), Positives = 24/44 (54%), Gaps = 1/44 (2%)
Query 93 RVNALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQEKKRL 136
R+N L N + K DK NYI E D++E+ RLK +K L
Sbjct 1236 RINDLMNFITQK-DKERNYIHHEEDKIEKNMLLRLKNNSTRKSL 1278
> hsa:9720 CCDC144A, FLJ43983, KIAA0565, MGC164650; coiled-coil
domain containing 144A
Length=1427
Score = 31.2 bits (69), Expect = 1.6, Method: Composition-based stats.
Identities = 23/68 (33%), Positives = 38/68 (55%), Gaps = 4/68 (5%)
Query 104 KLDKSINYITKELDEMEREEFFRLKKIQEKKRLAKEAEQKALEEAGKALKASGLLS-GTI 162
+L + + E++ +E EEF LKK E +L K+ E++ + + + SG L+ GT
Sbjct 494 ELQQDMQKFKNEVNTLE-EEFLALKK--EDVQLHKDVEEEMEKHRSNSTELSGTLTDGTT 550
Query 163 LGEDDDDL 170
+G DDD L
Sbjct 551 VGNDDDGL 558
> hsa:9851 MGC130040, MGC130041; KIAA0753
Length=967
Score = 30.4 bits (67), Expect = 2.7, Method: Composition-based stats.
Identities = 16/60 (26%), Positives = 34/60 (56%), Gaps = 1/60 (1%)
Query 88 KMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQ-EKKRLAKEAEQKALE 146
++ + ++++ L LD + TKEL+E++ EE +RL+++ LA + E+ L+
Sbjct 627 ELKAKEIDSMQKQRLDWLDAETSRRTKELNELKAEEMYRLQQLSVSATHLADKVEEAVLD 686
> pfa:PF07_0086 conserved Plasmodium membrane protein, unknown
function
Length=3429
Score = 30.4 bits (67), Expect = 2.8, Method: Composition-based stats.
Identities = 13/40 (32%), Positives = 25/40 (62%), Gaps = 1/40 (2%)
Query 83 LDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMERE 122
++ E+ N VN +N V K+D+ +N + KE+++M +E
Sbjct 795 MNEEVNKMNEEVNKMNKEV-NKMDEEVNKMNKEVNKMNKE 833
> pfa:PF11_0512 RESA-like protein with PHIST and DnaJ domains
Length=803
Score = 30.4 bits (67), Expect = 2.8, Method: Composition-based stats.
Identities = 14/56 (25%), Positives = 31/56 (55%), Gaps = 0/56 (0%)
Query 92 RRVNALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQEKKRLAKEAEQKALEE 147
RR+ LN +V L I Y +++ + F R+ +++ ++ A++A Q+ +E+
Sbjct 272 RRLTVLNQIVWKALSNQIEYSCRKIMTSDITSFIRINELEIMEQRAEKAAQEEMEK 327
> dre:558475 si:ch211-57h10.1
Length=239
Score = 30.4 bits (67), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 119 MEREEFFRLKKIQEKKRLAKEAEQKALEEAGKALK 153
+EREEF K E RLA ++QK L GK LK
Sbjct 54 LEREEFETRKAAAEASRLASGSQQKKLASTGKELK 88
> pfa:PFI1480w conserved Plasmodium protein, unknown function
Length=915
Score = 30.4 bits (67), Expect = 3.4, Method: Composition-based stats.
Identities = 25/66 (37%), Positives = 37/66 (56%), Gaps = 6/66 (9%)
Query 91 NRRVN-ALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQEKKRLA--KEAEQKALEE 147
N +N A+NNVV DK +N K+ D+ +E R+K+ +EKK++ K EQ LEE
Sbjct 452 NENINDAVNNVV--SEDKELNS-DKQSDKYISDELKRIKREEEKKKVMNWKSEEQTLLEE 508
Query 148 AGKALK 153
+ K
Sbjct 509 GLRIYK 514
> xla:398105 incenp-a, xl-incenp; inner centromere protein antigens
135/155kDa; K11515 inner centromere protein
Length=873
Score = 30.0 bits (66), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 27/41 (65%), Gaps = 0/41 (0%)
Query 114 KELDEMEREEFFRLKKIQEKKRLAKEAEQKALEEAGKALKA 154
K+ ++ ER E RL+K QE KRL +E ++KA E+A A A
Sbjct 701 KKREQQERLEQERLRKEQEAKRLQEEEQRKAKEQAAVAASA 741
> mmu:74477 4933427D14Rik, C85113, Kiaa0753; RIKEN cDNA 4933427D14
gene
Length=959
Score = 30.0 bits (66), Expect = 4.2, Method: Composition-based stats.
Identities = 16/35 (45%), Positives = 23/35 (65%), Gaps = 1/35 (2%)
Query 113 TKELDEMEREEFFRLKKIQ-EKKRLAKEAEQKALE 146
TKELDE++ EE RL+K+ +LA + E+ LE
Sbjct 643 TKELDELKAEEMDRLQKLSVSATQLADKVEEAVLE 677
> sce:YDL070W BDF2; Bdf2p; K11684 bromodomain-containing factor
1
Length=638
Score = 30.0 bits (66), Expect = 4.5, Method: Composition-based stats.
Identities = 17/62 (27%), Positives = 35/62 (56%), Gaps = 1/62 (1%)
Query 95 NALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQEKKRLAKE-AEQKALEEAGKALK 153
N + N + L++ + + EL +++R+E +L K +++K L K +KA++ + LK
Sbjct 462 NDITNPAIQYLEQKLKKMEVELQQLKRQELSKLSKERKRKHLGKTLLRRKAMKHSVDDLK 521
Query 154 AS 155
S
Sbjct 522 KS 523
> bbo:BBOV_III005600 17.m07500; Spherical Body Protein 2 truncated
copy 1 (SBP2)
Length=283
Score = 29.6 bits (65), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 25/76 (32%), Positives = 35/76 (46%), Gaps = 3/76 (3%)
Query 94 VNALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQEKKR---LAKEAEQKALEEAGK 150
V ALN+ + + DK KELDEM ++ + +K QE K+ KE E K +E K
Sbjct 177 VKALNDRIEAEQDKIREKTNKELDEMRNKDLKKEQKEQESKKEQESKKEQESKKEQEGQK 236
Query 151 ALKASGLLSGTILGED 166
+ G ED
Sbjct 237 EQEGQKEQEGQKEQED 252
> tgo:TGME49_015080 hypothetical protein
Length=3347
Score = 29.6 bits (65), Expect = 5.1, Method: Composition-based stats.
Identities = 26/112 (23%), Positives = 46/112 (41%), Gaps = 15/112 (13%)
Query 42 DILKNINLSAGGHVILAARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVV 101
D L++ S + D + LA + +A+ Q + F L ++ L N V
Sbjct 2909 DALESRERSDASEEMATVNDAEKRRLASQLLVATGQKSLFLLRPRFELVEETEGLLQNEV 2968
Query 102 LPKLDKSINYITKELDEMEREEFFRLKKIQEKKRLAKEAEQKALEEAGKALK 153
PK+DK + + L+ QEK+ L KE + +EE + ++
Sbjct 2969 GPKMDKEVEQV--------------LETNQEKESLRKETTGE-VEETNRGVR 3005
> hsa:255119 C4orf22, MGC35043; chromosome 4 open reading frame
22
Length=233
Score = 29.3 bits (64), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 30/51 (58%), Gaps = 0/51 (0%)
Query 117 DEMEREEFFRLKKIQEKKRLAKEAEQKALEEAGKALKASGLLSGTILGEDD 167
+ ++RE+F K E RLA+ A+QK L AGK L+ + L + + ED+
Sbjct 55 ERVKREDFEARKAAIEIARLAERAQQKTLTSAGKDLQDNFLTALAMREEDN 105
> ath:AT3G19515 hypothetical protein
Length=805
Score = 29.3 bits (64), Expect = 7.2, Method: Composition-based stats.
Identities = 18/56 (32%), Positives = 27/56 (48%), Gaps = 0/56 (0%)
Query 95 NALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQEKKRLAKEAEQKALEEAGK 150
N+LN ++P DKS+ ++ E +M E L K EK + E + E GK
Sbjct 724 NSLNEHIMPDSDKSLGNVSGEEIKMVTEGLTSLSKFGEKGPQERMHEPVEIVERGK 779
> tgo:TGME49_003810 kelch motif domain-containing protein
Length=531
Score = 29.3 bits (64), Expect = 7.4, Method: Composition-based stats.
Identities = 17/70 (24%), Positives = 34/70 (48%), Gaps = 0/70 (0%)
Query 67 LAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFR 126
L V ++L+ LD +K+T +A L ++ +I+ +T+E+ + E
Sbjct 459 LVTFVSFSALRLQVEALDEIVKLTTETQSAEKVAKLAQMQNAISKLTEEMTAIREENESL 518
Query 127 LKKIQEKKRL 136
K++ E +RL
Sbjct 519 KKRLSEVERL 528
> ath:AT4G25690 hypothetical protein
Length=191
Score = 28.9 bits (63), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 22/81 (27%), Positives = 41/81 (50%), Gaps = 21/81 (25%)
Query 109 INYITKELDEMEREEFFRLKKIQEKKRLAKEA--------------------EQKALEEA 148
+ +T+++ + ++EF R+K+ ++KKR ++A EQ+ L+E
Sbjct 10 VQPVTRKVKKHGKDEFDRIKQAEKKKRRLEKALATSAAIRAELEKKKQKRLEEQQRLDEE 69
Query 149 GKALKASGLLSGTILGEDDDD 169
G A+ A + +LGED DD
Sbjct 70 GAAI-AEAVALHVLLGEDSDD 89
> cel:Y102A11A.2 hypothetical protein
Length=772
Score = 28.9 bits (63), Expect = 8.3, Method: Composition-based stats.
Identities = 22/67 (32%), Positives = 34/67 (50%), Gaps = 12/67 (17%)
Query 114 KELDEMEREEFFRLKKIQEKKRLAKEAEQKALEEAGK-----------ALKASGL-LSGT 161
K+ +E R + KK +++K AK EQ+ +EEA K L+A L L T
Sbjct 483 KKQEEEIRAKLEAKKKAEQEKERAKREEQRRIEEAAKLEYRRRIEESQRLEAERLALEAT 542
Query 162 ILGEDDD 168
++ ED+D
Sbjct 543 MVDEDED 549
> cel:C33E10.6 hypothetical protein
Length=1014
Score = 28.9 bits (63), Expect = 8.5, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 35/65 (53%), Gaps = 10/65 (15%)
Query 84 DSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQE--KKRLAKEAE 141
+SE++ ++ VN +NN ++ L K IN KEL E F L + E K+ ++ + E
Sbjct 591 NSEVQNKDKEVNEINNKIIQHLKKIIN--AKELGE------FTLPMLNECIKEVISNDNE 642
Query 142 QKALE 146
K +E
Sbjct 643 NKTIE 647
> dre:558350 eme1, fc30c07, wu:fc30c07; essential meiotic endonuclease
1 homolog 1 (S. pombe); K10882 crossover junction endonuclease
EME1 [EC:3.1.22.-]
Length=556
Score = 28.9 bits (63), Expect = 8.8, Method: Composition-based stats.
Identities = 24/95 (25%), Positives = 43/95 (45%), Gaps = 2/95 (2%)
Query 72 KLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQ 131
K++S T ++ + + + A N L K K+ + E R+ R + +
Sbjct 140 KISSNITRHYSSNHDAPYLTKEPEAENGTYLAKRKKTPAEVEAARQEALRKRAIREHQQE 199
Query 132 EKKRLAKEAEQKALEEAGKALKASGLLSGTILGED 166
E+ RL E+KAL +A KAL+ + T++ D
Sbjct 200 ERGRL--RMEKKALADAVKALRPEECIKHTVVTVD 232
> ath:AT1G20760 calcium-binding EF hand family protein
Length=1019
Score = 28.9 bits (63), Expect = 9.4, Method: Composition-based stats.
Identities = 28/94 (29%), Positives = 47/94 (50%), Gaps = 6/94 (6%)
Query 58 AARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPK--LDKSINYITKE 115
+A EA A+ K+ Q A+ +DS K+ R + ++VL K D +N I++
Sbjct 526 SASSNLPEAAADEEKVDEKQNAY--MDSREKLDYYRTK-MQDIVLYKSRCDNRLNEISER 582
Query 116 LDEMEREEFFRLKKIQEK-KRLAKEAEQKALEEA 148
+RE KK +EK K++A+ + +EEA
Sbjct 583 ASADKREAETLAKKYEEKYKQVAEIGSKLTIEEA 616
Lambda K H
0.318 0.136 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4341553636
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40