bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3590_orf1
Length=178
Score E
Sequences producing significant alignments: (Bits) Value
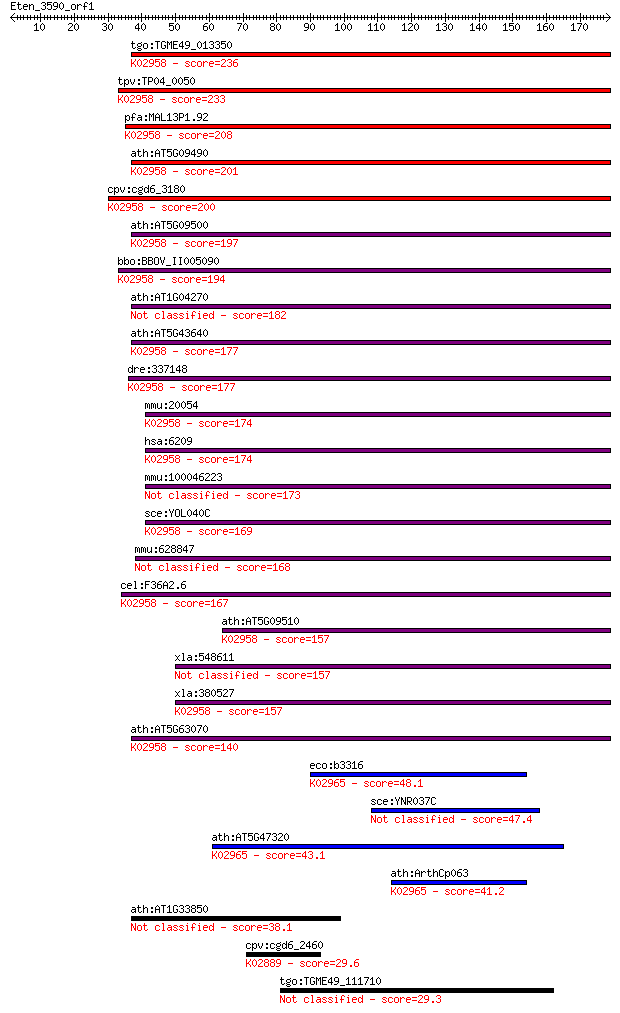
tgo:TGME49_013350 40S ribosomal protein S15, putative ; K02958... 236 4e-62
tpv:TP04_0050 40S ribosomal protein S15; K02958 small subunit ... 233 2e-61
pfa:MAL13P1.92 40S ribosomal protein S15/S19, putative; K02958... 208 7e-54
ath:AT5G09490 40S ribosomal protein S15 (RPS15B); K02958 small... 201 1e-51
cpv:cgd6_3180 40S ribosomal protein S15 ; K02958 small subunit... 200 2e-51
ath:AT5G09500 40S ribosomal protein S15 (RPS15C); K02958 small... 197 2e-50
bbo:BBOV_II005090 18.m06424; 40S ribosomal protein S15; K02958... 194 1e-49
ath:AT1G04270 RPS15; RPS15 (CYTOSOLIC RIBOSOMAL PROTEIN S15); ... 182 5e-46
ath:AT5G43640 40S ribosomal protein S15 (RPS15E); K02958 small... 177 2e-44
dre:337148 rps15, MGC103714, wu:fa93f04, wu:fa99b07, zgc:10371... 177 2e-44
mmu:20054 Rps15, rig; ribosomal protein S15; K02958 small subu... 174 1e-43
hsa:6209 RPS15, MGC111130, RIG; ribosomal protein S15; K02958 ... 174 1e-43
mmu:100046223 40S ribosomal protein S15-like 173 2e-43
sce:YOL040C RPS15, RPS21; Rps15p; K02958 small subunit ribosom... 169 3e-42
mmu:628847 Gm6921, EG628847; predicted pseudogene 6921 168 9e-42
cel:F36A2.6 rps-15; Ribosomal Protein, Small subunit family me... 167 1e-41
ath:AT5G09510 40S ribosomal protein S15 (RPS15D); K02958 small... 157 1e-38
xla:548611 rps15; 40S ribosomal protein S15 157 1e-38
xla:380527 rps15, MGC64489, rig; ribosomal protein S15; K02958... 157 1e-38
ath:AT5G63070 40S ribosomal protein S15, putative; K02958 smal... 140 2e-33
eco:b3316 rpsS, ECK3303, JW3278; 30S ribosomal subunit protein... 48.1 2e-05
sce:YNR037C RSM19; Mitochondrial ribosomal protein of the smal... 47.4 3e-05
ath:AT5G47320 RPS19; RPS19 (RIBOSOMAL PROTEIN S19); RNA bindin... 43.1 5e-04
ath:ArthCp063 rps19; 30S ribosomal protein S19; K02965 small s... 41.2 0.002
ath:AT1G33850 40S ribosomal protein S15, putative 38.1 0.017
cpv:cgd6_2460 60s ribosomal protein L21 ; K02889 large subunit... 29.6 6.0
tgo:TGME49_111710 hypothetical protein 29.3 6.5
> tgo:TGME49_013350 40S ribosomal protein S15, putative ; K02958
small subunit ribosomal protein S15e
Length=150
Score = 236 bits (601), Expect = 4e-62, Method: Compositional matrix adjust.
Identities = 111/142 (78%), Positives = 129/142 (90%), Gaps = 0/142 (0%)
Query 37 ASVPRKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMNKLRNA 96
A+ P++RTFRTFSYRGV+L+KL+ + ++ L++LFRARQRRKF RGI +KA LM KLR +
Sbjct 9 ANQPKRRTFRTFSYRGVELDKLLDMKMEDLLELFRARQRRKFQRGIKRKATTLMKKLRIS 68
Query 97 KKDLPYGEKPPPVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFSISY 156
KK+ +GEKP PVNTHLR++VVVPEMIGS+VGVYNGKQFINVEIKPEMVGYYLGEFSISY
Sbjct 69 KKNCAFGEKPEPVNTHLRDLVVVPEMIGSVVGVYNGKQFINVEIKPEMVGYYLGEFSISY 128
Query 157 KPVRHGKPGIGATHSSRFIPLK 178
KPVRHGKPGIGATHSSRFIPLK
Sbjct 129 KPVRHGKPGIGATHSSRFIPLK 150
> tpv:TP04_0050 40S ribosomal protein S15; K02958 small subunit
ribosomal protein S15e
Length=148
Score = 233 bits (594), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 111/148 (75%), Positives = 130/148 (87%), Gaps = 2/148 (1%)
Query 33 MAEDASVP--RKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLM 90
M E+ V +KRTFRTF YRG +LEKL+ +P+DKL +L ARQRR+FSRG+ +++L L+
Sbjct 1 MVEEQGVQILKKRTFRTFKYRGYELEKLLEMPIDKLTELLPARQRRRFSRGVKRQSLTLL 60
Query 91 NKLRNAKKDLPYGEKPPPVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLG 150
KLR AKKDLPYG+KP PV THLRN+VV+PEMIGSIVGV+NGKQ+INVEIKPEMVGYYLG
Sbjct 61 KKLRAAKKDLPYGQKPEPVKTHLRNMVVIPEMIGSIVGVHNGKQYINVEIKPEMVGYYLG 120
Query 151 EFSISYKPVRHGKPGIGATHSSRFIPLK 178
EFSI+YKPVRHGKPGIGATHSSRFIPLK
Sbjct 121 EFSITYKPVRHGKPGIGATHSSRFIPLK 148
> pfa:MAL13P1.92 40S ribosomal protein S15/S19, putative; K02958
small subunit ribosomal protein S15e
Length=145
Score = 208 bits (530), Expect = 7e-54, Method: Compositional matrix adjust.
Identities = 102/144 (70%), Positives = 123/144 (85%), Gaps = 0/144 (0%)
Query 35 EDASVPRKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMNKLR 94
EDA+ P+KRTFRTF YRGVDL+KL+ L D+L+ LF+ARQRRKF RGI++KA L+ K+R
Sbjct 2 EDANKPKKRTFRTFQYRGVDLDKLLDLSQDELIKLFKARQRRKFQRGISKKAKSLLKKIR 61
Query 95 NAKKDLPYGEKPPPVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFSI 154
+KK+ GEKP PV THLRN+ ++PEM+GSIV V+NGKQ+ NVEIKPEM+GYYLGEFSI
Sbjct 62 KSKKNCEPGEKPNPVPTHLRNMTIIPEMVGSIVAVHNGKQYTNVEIKPEMIGYYLGEFSI 121
Query 155 SYKPVRHGKPGIGATHSSRFIPLK 178
+YK RHGKPGIGATHSSRFIPLK
Sbjct 122 TYKHTRHGKPGIGATHSSRFIPLK 145
> ath:AT5G09490 40S ribosomal protein S15 (RPS15B); K02958 small
subunit ribosomal protein S15e
Length=152
Score = 201 bits (510), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 93/142 (65%), Positives = 117/142 (82%), Gaps = 0/142 (0%)
Query 37 ASVPRKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMNKLRNA 96
A++ +KRTF+ FS++GVDL+ L+ +P D LV+LF +R RR+ SRG+ +K + L+ KLR A
Sbjct 11 AALAKKRTFKKFSFKGVDLDALLDMPTDDLVELFPSRIRRRMSRGLTRKPMALIKKLRKA 70
Query 97 KKDLPYGEKPPPVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFSISY 156
K D P GEKP V THLRN+V++PEMIGSI+GVYNGK F +EIKPEM+G+YL EFSI+Y
Sbjct 71 KLDAPAGEKPEVVRTHLRNMVIMPEMIGSIIGVYNGKTFNQIEIKPEMIGHYLAEFSITY 130
Query 157 KPVRHGKPGIGATHSSRFIPLK 178
KPVRHGKPG GATHSSRFIPLK
Sbjct 131 KPVRHGKPGHGATHSSRFIPLK 152
> cpv:cgd6_3180 40S ribosomal protein S15 ; K02958 small subunit
ribosomal protein S15e
Length=152
Score = 200 bits (509), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 93/150 (62%), Positives = 122/150 (81%), Gaps = 3/150 (2%)
Query 30 QNKMAEDASVPRKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRG-INQKALP 88
+ KMA+ +KRTFRT+SYRGVDL+KL+T+ LD++V+L AR+RRK +RG +N++
Sbjct 5 EKKMADTEQ--KKRTFRTYSYRGVDLDKLLTMKLDEVVELLPARKRRKIARGCLNRRTAA 62
Query 89 LMNKLRNAKKDLPYGEKPPPVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYY 148
+ KLR +K + P GEKP V THLRN+V++PEM+GS+ GVYNGK ++ VEIKPEM+G Y
Sbjct 63 FIAKLRKSKAECPMGEKPVAVRTHLRNMVILPEMVGSVAGVYNGKTYVTVEIKPEMIGMY 122
Query 149 LGEFSISYKPVRHGKPGIGATHSSRFIPLK 178
LGEFSI+YKPVRHGKPG+G+T SSRFIPLK
Sbjct 123 LGEFSITYKPVRHGKPGVGSTSSSRFIPLK 152
> ath:AT5G09500 40S ribosomal protein S15 (RPS15C); K02958 small
subunit ribosomal protein S15e
Length=150
Score = 197 bits (500), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 91/142 (64%), Positives = 116/142 (81%), Gaps = 0/142 (0%)
Query 37 ASVPRKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMNKLRNA 96
A + +KRTF+ FS+RGVDL+ L+ + D LV LF +R RR+FSRG+ +K + L+ KLR A
Sbjct 9 AGIVKKRTFKKFSFRGVDLDALLDMSTDDLVKLFPSRIRRRFSRGLTRKPMALIKKLRKA 68
Query 97 KKDLPYGEKPPPVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFSISY 156
K + P GEKP V THLRN+++VPEMIGS++GVYNGK F VEIKPEM+G+YL EFSISY
Sbjct 69 KIEAPAGEKPAAVRTHLRNMIIVPEMIGSVIGVYNGKTFNQVEIKPEMIGHYLAEFSISY 128
Query 157 KPVRHGKPGIGATHSSRFIPLK 178
KPV+HG+PG+GAT+SSRFIPLK
Sbjct 129 KPVKHGRPGVGATNSSRFIPLK 150
> bbo:BBOV_II005090 18.m06424; 40S ribosomal protein S15; K02958
small subunit ribosomal protein S15e
Length=147
Score = 194 bits (494), Expect = 1e-49, Method: Compositional matrix adjust.
Identities = 101/147 (68%), Positives = 124/147 (84%), Gaps = 1/147 (0%)
Query 33 MAEDASVP-RKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMN 91
MA+ A+ RKRTFRTF +RG +LEKL+ +P++ V L ++RQRR+FSRGI + L+
Sbjct 1 MADQAAQGNRKRTFRTFKFRGYELEKLLEMPMENFVGLLKSRQRRRFSRGIKTRIRTLLK 60
Query 92 KLRNAKKDLPYGEKPPPVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGE 151
KL+ AKKDL YGEKP PV THLR+ +++PEMIGSIVGVYNGKQ+INVEIKPEMVG+Y+GE
Sbjct 61 KLKAAKKDLAYGEKPEPVKTHLRDTIIIPEMIGSIVGVYNGKQYINVEIKPEMVGHYVGE 120
Query 152 FSISYKPVRHGKPGIGATHSSRFIPLK 178
FSI+YKPV HGKPGIG+THSSRFIPLK
Sbjct 121 FSITYKPVMHGKPGIGSTHSSRFIPLK 147
> ath:AT1G04270 RPS15; RPS15 (CYTOSOLIC RIBOSOMAL PROTEIN S15);
structural constituent of ribosome
Length=151
Score = 182 bits (462), Expect = 5e-46, Method: Compositional matrix adjust.
Identities = 93/142 (65%), Positives = 118/142 (83%), Gaps = 1/142 (0%)
Query 37 ASVPRKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMNKLRNA 96
A VP+KRTF+ F+++GVDL+ L+ + D LV LF +R RR+FSRG+ +K + L+ KLR
Sbjct 11 AGVPKKRTFKKFAFKGVDLDALLDMSTDDLVKLFSSRIRRRFSRGLTRKPMALIKKLRK- 69
Query 97 KKDLPYGEKPPPVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFSISY 156
K++ P GEKP PV THLRN+++VPEMIGSI+GVYNGK F VEIKPEM+G+YL EFSISY
Sbjct 70 KREAPQGEKPEPVRTHLRNMIIVPEMIGSIIGVYNGKTFNQVEIKPEMIGHYLAEFSISY 129
Query 157 KPVRHGKPGIGATHSSRFIPLK 178
KPV+HG+PG+GATHSSRFIPLK
Sbjct 130 KPVKHGRPGVGATHSSRFIPLK 151
> ath:AT5G43640 40S ribosomal protein S15 (RPS15E); K02958 small
subunit ribosomal protein S15e
Length=149
Score = 177 bits (449), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 90/142 (63%), Positives = 116/142 (81%), Gaps = 0/142 (0%)
Query 37 ASVPRKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMNKLRNA 96
A + +KRTF+ FS+RGVDL+ L+ + ++ LV F +R RR+FSRG+ +K + L+ KLR A
Sbjct 8 AGIVKKRTFKKFSFRGVDLDALLDMSIEDLVKHFSSRIRRRFSRGLTRKPMALIKKLRKA 67
Query 97 KKDLPYGEKPPPVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFSISY 156
K + P GEKP V THLRN+++VPEMIGSI+GVYNGK F VEIKPEM+G+YL EFSISY
Sbjct 68 KMEAPAGEKPASVRTHLRNMIIVPEMIGSIIGVYNGKTFNQVEIKPEMIGHYLAEFSISY 127
Query 157 KPVRHGKPGIGATHSSRFIPLK 178
KPV+HG+PG+GAT+SSRFIPLK
Sbjct 128 KPVKHGRPGVGATNSSRFIPLK 149
> dre:337148 rps15, MGC103714, wu:fa93f04, wu:fa99b07, zgc:103714,
zgc:92743; ribosomal protein S15; K02958 small subunit
ribosomal protein S15e
Length=145
Score = 177 bits (448), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 89/143 (62%), Positives = 117/143 (81%), Gaps = 0/143 (0%)
Query 36 DASVPRKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMNKLRN 95
D +KRTFR F+YRGVDL++L+ + ++L+ L+ ARQRR+ +RG+ +K L+ +LR
Sbjct 3 DTEQKKKRTFRKFTYRGVDLDQLLDMSYEQLMQLYSARQRRRLNRGLRRKQQSLLKRLRK 62
Query 96 AKKDLPYGEKPPPVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFSIS 155
AKK+ P EKP V THLR++V++PEM+GS+VGVYNGK F VEIKPEM+G+YLGEFSI+
Sbjct 63 AKKEAPPMEKPEVVKTHLRDMVILPEMVGSMVGVYNGKTFNQVEIKPEMIGHYLGEFSIT 122
Query 156 YKPVRHGKPGIGATHSSRFIPLK 178
YKPV+HG+PGIGATHSSRFIPLK
Sbjct 123 YKPVKHGRPGIGATHSSRFIPLK 145
> mmu:20054 Rps15, rig; ribosomal protein S15; K02958 small subunit
ribosomal protein S15e
Length=145
Score = 174 bits (442), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 87/138 (63%), Positives = 116/138 (84%), Gaps = 0/138 (0%)
Query 41 RKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMNKLRNAKKDL 100
+KRTFR F+YRGVDL++L+ + ++L+ L+ ARQRR+ +RG+ +K L+ +LR AKK+
Sbjct 8 KKRTFRKFTYRGVDLDQLLDMSYEQLMQLYSARQRRRLNRGLRRKQHSLLKRLRKAKKEA 67
Query 101 PYGEKPPPVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFSISYKPVR 160
P EKP V THLR+++++PEM+GS+VGVYNGK F VEIKPEM+G+YLGEFSI+YKPV+
Sbjct 68 PPMEKPEVVKTHLRDMIILPEMVGSMVGVYNGKTFNQVEIKPEMIGHYLGEFSITYKPVK 127
Query 161 HGKPGIGATHSSRFIPLK 178
HG+PGIGATHSSRFIPLK
Sbjct 128 HGRPGIGATHSSRFIPLK 145
> hsa:6209 RPS15, MGC111130, RIG; ribosomal protein S15; K02958
small subunit ribosomal protein S15e
Length=145
Score = 174 bits (442), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 87/138 (63%), Positives = 116/138 (84%), Gaps = 0/138 (0%)
Query 41 RKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMNKLRNAKKDL 100
+KRTFR F+YRGVDL++L+ + ++L+ L+ ARQRR+ +RG+ +K L+ +LR AKK+
Sbjct 8 KKRTFRKFTYRGVDLDQLLDMSYEQLMQLYSARQRRRLNRGLRRKQHSLLKRLRKAKKEA 67
Query 101 PYGEKPPPVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFSISYKPVR 160
P EKP V THLR+++++PEM+GS+VGVYNGK F VEIKPEM+G+YLGEFSI+YKPV+
Sbjct 68 PPMEKPEVVKTHLRDMIILPEMVGSMVGVYNGKTFNQVEIKPEMIGHYLGEFSITYKPVK 127
Query 161 HGKPGIGATHSSRFIPLK 178
HG+PGIGATHSSRFIPLK
Sbjct 128 HGRPGIGATHSSRFIPLK 145
> mmu:100046223 40S ribosomal protein S15-like
Length=145
Score = 173 bits (439), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 86/138 (62%), Positives = 116/138 (84%), Gaps = 0/138 (0%)
Query 41 RKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMNKLRNAKKDL 100
+KRTFR F+YRGVDL++L+ + ++L+ L+ ARQRR+ +RG+ +K L+ +LR AKK+
Sbjct 8 KKRTFRKFTYRGVDLDQLLDMSYEQLMQLYSARQRRRLNRGLRRKQHSLLKRLRKAKKEA 67
Query 101 PYGEKPPPVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFSISYKPVR 160
P EKP V THLR+++++PEM+GS+VG+YNGK F VEIKPEM+G+YLGEFSI+YKPV+
Sbjct 68 PPMEKPEVVKTHLRDMIILPEMVGSMVGMYNGKTFNQVEIKPEMIGHYLGEFSITYKPVK 127
Query 161 HGKPGIGATHSSRFIPLK 178
HG+PGIGATHSSRFIPLK
Sbjct 128 HGRPGIGATHSSRFIPLK 145
> sce:YOL040C RPS15, RPS21; Rps15p; K02958 small subunit ribosomal
protein S15e
Length=142
Score = 169 bits (429), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 82/138 (59%), Positives = 102/138 (73%), Gaps = 3/138 (2%)
Query 41 RKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMNKLRNAKKDL 100
+KR F+T SYRGVDLEKL+ + + V L AR RR+F+RG+ K M KLR AK
Sbjct 8 KKRVFKTHSYRGVDLEKLLEMSTEDFVKLAPARVRRRFARGMTSKPAGFMKKLRAAKLAA 67
Query 101 PYGEKPPPVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFSISYKPVR 160
P EKP PV TH+RN+++VPEMIGS+VG+YNGK F VEI+PEM+G+YLGEFSI+Y PVR
Sbjct 68 PENEKPAPVRTHMRNMIIVPEMIGSVVGIYNGKAFNQVEIRPEMLGHYLGEFSITYTPVR 127
Query 161 HGKPGIGATHSSRFIPLK 178
HG+ G +SRFIPLK
Sbjct 128 HGRAG---ATTSRFIPLK 142
> mmu:628847 Gm6921, EG628847; predicted pseudogene 6921
Length=148
Score = 168 bits (425), Expect = 9e-42, Method: Compositional matrix adjust.
Identities = 75/141 (53%), Positives = 112/141 (79%), Gaps = 0/141 (0%)
Query 38 SVPRKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMNKLRNAK 97
++ +K+TF F YRGVDL++L+ + ++L+ L+ A+QR+ +RG+ K ++ +LR +K
Sbjct 8 NLKKKQTFCKFIYRGVDLDQLLHMSYEQLIQLYSAQQRQCLNRGLRLKQHSMLKRLRKSK 67
Query 98 KDLPYGEKPPPVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFSISYK 157
K+ P EKP V T+LR+++++PEM+GS+VGVYN K F VEIKP+M+G+YLGEFSI++K
Sbjct 68 KEAPPMEKPEVVKTNLRDMIILPEMVGSVVGVYNSKTFNQVEIKPKMIGHYLGEFSITFK 127
Query 158 PVRHGKPGIGATHSSRFIPLK 178
PV+ G+PG+GATHSSRFIPLK
Sbjct 128 PVKQGRPGMGATHSSRFIPLK 148
> cel:F36A2.6 rps-15; Ribosomal Protein, Small subunit family
member (rps-15); K02958 small subunit ribosomal protein S15e
Length=151
Score = 167 bits (424), Expect = 1e-41, Method: Compositional matrix adjust.
Identities = 80/145 (55%), Positives = 109/145 (75%), Gaps = 0/145 (0%)
Query 34 AEDASVPRKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMNKL 93
A A + +KRTFR F YRGVDL++L+ + ++ L R RR+ RG+ +K L L+ K+
Sbjct 7 AHLAELKKKRTFRKFMYRGVDLDQLLDMSREQFTKLLPCRMRRRLDRGLKRKHLALIAKV 66
Query 94 RNAKKDLPYGEKPPPVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFS 153
+ AKK EKP V THLR+++++PE++G ++G+YNGK F EIKPEM+G+YLGEF+
Sbjct 67 QKAKKAAGVLEKPATVKTHLRDMIILPELVGGVIGIYNGKVFNQTEIKPEMIGFYLGEFA 126
Query 154 ISYKPVRHGKPGIGATHSSRFIPLK 178
ISYKPV+HG+PGIGATHSSRFIPLK
Sbjct 127 ISYKPVKHGRPGIGATHSSRFIPLK 151
> ath:AT5G09510 40S ribosomal protein S15 (RPS15D); K02958 small
subunit ribosomal protein S15e
Length=118
Score = 157 bits (398), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 81/115 (70%), Positives = 98/115 (85%), Gaps = 0/115 (0%)
Query 64 DKLVDLFRARQRRKFSRGINQKALPLMNKLRNAKKDLPYGEKPPPVNTHLRNVVVVPEMI 123
D LV LF +R RR+FSRG+ +K + L+ KLR AK++ P GEKP PV THLRN+++VPEMI
Sbjct 4 DDLVKLFSSRIRRRFSRGLTRKPMALIKKLRKAKREAPAGEKPEPVRTHLRNMIIVPEMI 63
Query 124 GSIVGVYNGKQFINVEIKPEMVGYYLGEFSISYKPVRHGKPGIGATHSSRFIPLK 178
GSI+GVYNGK F VEIKPEM+G+YL EFSISYKPV+HG+PG+GATHSSRFIPLK
Sbjct 64 GSIIGVYNGKTFNQVEIKPEMIGHYLAEFSISYKPVKHGRPGVGATHSSRFIPLK 118
> xla:548611 rps15; 40S ribosomal protein S15
Length=145
Score = 157 bits (398), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 79/129 (61%), Positives = 108/129 (83%), Gaps = 0/129 (0%)
Query 50 YRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMNKLRNAKKDLPYGEKPPPV 109
YRGVDL++L+ + ++++ L+ ARQRR+ +RG+ +K L+ +LR AKK+ P EKP +
Sbjct 17 YRGVDLDQLLDMSYEQVMQLYCARQRRRLNRGLRRKQNSLLKRLRKAKKEAPPMEKPEVI 76
Query 110 NTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFSISYKPVRHGKPGIGAT 169
THLR+++++PEM+GS+VGVYNGK F VEIKPEM+G+YLGEFSI+YKPV+HG+PGIGAT
Sbjct 77 KTHLRDMIILPEMVGSMVGVYNGKAFNQVEIKPEMIGHYLGEFSITYKPVKHGRPGIGAT 136
Query 170 HSSRFIPLK 178
HSSRFIPLK
Sbjct 137 HSSRFIPLK 145
> xla:380527 rps15, MGC64489, rig; ribosomal protein S15; K02958
small subunit ribosomal protein S15e
Length=145
Score = 157 bits (398), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 79/129 (61%), Positives = 108/129 (83%), Gaps = 0/129 (0%)
Query 50 YRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMNKLRNAKKDLPYGEKPPPV 109
YRGVDL++L+ + ++++ L+ ARQRR+ +RG+ +K L+ +LR AKK+ P EKP +
Sbjct 17 YRGVDLDQLLDMSYEQVMQLYCARQRRRLNRGLRRKQNSLLKRLRKAKKEAPPMEKPEVI 76
Query 110 NTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFSISYKPVRHGKPGIGAT 169
THLR+++++PEM+GS+VGVYNGK F VEIKPEM+G+YLGEFSI+YKPV+HG+PGIGAT
Sbjct 77 KTHLRDMIILPEMVGSMVGVYNGKAFNQVEIKPEMIGHYLGEFSITYKPVKHGRPGIGAT 136
Query 170 HSSRFIPLK 178
HSSRFIPLK
Sbjct 137 HSSRFIPLK 145
> ath:AT5G63070 40S ribosomal protein S15, putative; K02958 small
subunit ribosomal protein S15e
Length=160
Score = 140 bits (353), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 72/148 (48%), Positives = 100/148 (67%), Gaps = 6/148 (4%)
Query 37 ASVPRKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMNKLRNA 96
A +KRTF+ FS+RG +++ L+ + L LF AR RR+F RG+ ++ L L+ KLR A
Sbjct 13 AVATKKRTFKKFSFRGFNVDALLKMSNVDLAKLFNARVRRRFYRGLKKQPLILIKKLRRA 72
Query 97 KKDLPYGEKPPP--VNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFSI 154
KK+ K P V THLRN+++VPEMIGS+VGV+NGK+F + IKPEM+G+YL EFS+
Sbjct 73 KKEASDENKMKPEVVKTHLRNMIIVPEMIGSVVGVHNGKKFNEIVIKPEMIGHYLAEFSM 132
Query 155 SYKPVRHGKPGIGAT----HSSRFIPLK 178
+ K V H +P I S+RFIPL+
Sbjct 133 TCKKVNHHRPRICGCCCFRRSTRFIPLR 160
> eco:b3316 rpsS, ECK3303, JW3278; 30S ribosomal subunit protein
S19; K02965 small subunit ribosomal protein S19
Length=92
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 39/64 (60%), Gaps = 1/64 (1%)
Query 90 MNKLRNAKKDLPYGEKPPPVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYL 149
++ L+ +K + G+K P + T R + P MIG + V+NG+Q + V + EMVG+ L
Sbjct 13 LHLLKKVEKAVESGDKKP-LRTWSRRSTIFPNMIGLTIAVHNGRQHVPVFVTDEMVGHKL 71
Query 150 GEFS 153
GEF+
Sbjct 72 GEFA 75
> sce:YNR037C RSM19; Mitochondrial ribosomal protein of the small
subunit, has similarity to E. coli S19 ribosomal protein
Length=91
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 34/50 (68%), Gaps = 0/50 (0%)
Query 108 PVNTHLRNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFSISYK 157
P+ T+ R ++P+ +G ++NGK+++ +EI +MVG+ LGEF+ + K
Sbjct 32 PIRTNARAATILPQFVGLKFQIHNGKEYVPIEISEDMVGHKLGEFAPTRK 81
> ath:AT5G47320 RPS19; RPS19 (RIBOSOMAL PROTEIN S19); RNA binding;
K02965 small subunit ribosomal protein S19
Length=212
Score = 43.1 bits (100), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 27/104 (25%), Positives = 48/104 (46%), Gaps = 7/104 (6%)
Query 61 LPLDKLVDLFRARQRRKFSRGINQKALPLMNKLRNAKKDLPYGEKPPPVNTHLRNVVVVP 120
L LD+ ++ ++ + SR + + P ++ KK+ K R ++P
Sbjct 115 LSLDESIEEAEKKENKMMSRSVWKD--PFVDAFLMKKKNAALNRKIWS-----RRSTILP 167
Query 121 EMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFSISYKPVRHGKP 164
E + S V +YNGK + +I VG+ GEF+ + K +H K
Sbjct 168 EYVDSAVRIYNGKTHVRCKITEGKVGHKFGEFAFTRKVKKHAKA 211
> ath:ArthCp063 rps19; 30S ribosomal protein S19; K02965 small
subunit ribosomal protein S19
Length=92
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/40 (42%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 114 RNVVVVPEMIGSIVGVYNGKQFINVEIKPEMVGYYLGEFS 153
R ++P MIG + ++NG++ + V I MVG+ LGEFS
Sbjct 36 RASTIIPTMIGHTIAIHNGREHLPVYIIDLMVGHKLGEFS 75
> ath:AT1G33850 40S ribosomal protein S15, putative
Length=70
Score = 38.1 bits (87), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 29/62 (46%), Positives = 42/62 (67%), Gaps = 0/62 (0%)
Query 37 ASVPRKRTFRTFSYRGVDLEKLVTLPLDKLVDLFRARQRRKFSRGINQKALPLMNKLRNA 96
A + +KR F+ FS+RGVDL+ L+ + + LV F +R RR+FSRG +K + L+ KLR A
Sbjct 8 AGIVKKRIFKKFSFRGVDLDALLDMSTEDLVKHFSSRIRRRFSRGFTRKPMALIKKLRKA 67
Query 97 KK 98
K
Sbjct 68 VK 69
> cpv:cgd6_2460 60s ribosomal protein L21 ; K02889 large subunit
ribosomal protein L21e
Length=159
Score = 29.6 bits (65), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 11/22 (50%), Positives = 17/22 (77%), Gaps = 0/22 (0%)
Query 71 RARQRRKFSRGINQKALPLMNK 92
RAR R KFS+G QK +P++++
Sbjct 8 RARTRSKFSKGFRQKGVPMLSR 29
> tgo:TGME49_111710 hypothetical protein
Length=628
Score = 29.3 bits (64), Expect = 6.5, Method: Composition-based stats.
Identities = 25/89 (28%), Positives = 37/89 (41%), Gaps = 10/89 (11%)
Query 81 GINQ---KALPLMNKLRNAKKDLPYGEKPPPVNTHLRNVVVVPEMIGSIVGVYN-----G 132
G+NQ + +M + A D +G PP N V V E + + V N G
Sbjct 219 GVNQVDAASAEVMKMIEAALGD--FGTAPPDANVESERSVSVEENLAAREDVVNVAHEIG 276
Query 133 KQFINVEIKPEMVGYYLGEFSISYKPVRH 161
K+F E+ + G + EF I V+H
Sbjct 277 KRFGVAELYANLAGAFCDEFLIPAGLVKH 305
Lambda K H
0.322 0.140 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4730349484
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40