bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3598_orf1
Length=174
Score E
Sequences producing significant alignments: (Bits) Value
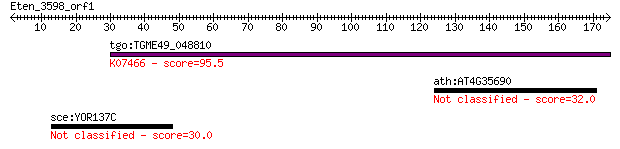
tgo:TGME49_048810 hypothetical protein ; K07466 replication fa... 95.5 8e-20
ath:AT4G35690 hypothetical protein 32.0 0.98
sce:YOR137C SIA1; Protein of unassigned function involved in a... 30.0 3.8
> tgo:TGME49_048810 hypothetical protein ; K07466 replication
factor A1
Length=497
Score = 95.5 bits (236), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 49/156 (31%), Positives = 85/156 (54%), Gaps = 11/156 (7%)
Query 30 AFTLNSGPYRNIPIYVHLAKGLPADAWEQVHTNPLFGEWAANLCHEGRLKADRITIQQVK 89
F+L + + + V L + WE++ ++ +F +W + RL+ R+ ++ +
Sbjct 14 GFSLTTQSHGECKVRVDLPSSISQADWERIFSSRMFQDWLSVYATADRLRLLRVALESSQ 73
Query 90 ----------PEICMNVEATTSQGTSVSGPVVLRPMQSAILIVLRNSVTNLELCVFCKRP 139
+ M VEA +G SGPV LRP + A+L++LRN+ T ++C+F K+P
Sbjct 74 HCKSSGEEKLTSVAMLVEAADKEGEVFSGPVYLRPQRRAVLVLLRNTETRTDMCLFVKKP 133
Query 140 ELATGLSESLGLVEGSFDAE-GKLEGPCAALLEKHL 174
L+ GL+++L L EG F+AE G+ GP A +E+ L
Sbjct 134 NLSVGLADTLELPEGEFEAETGRFVGPAAEEVERQL 169
> ath:AT4G35690 hypothetical protein
Length=284
Score = 32.0 bits (71), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 26/48 (54%), Gaps = 5/48 (10%)
Query 124 RNSVTNLELCVFCKRPELATGLSESLGLVEGSFDA-EGKLEGPCAALL 170
+N + NL+L +FC R +L L E VE S D E KLEG L+
Sbjct 229 KNELENLDLEIFCSRNDLQKKLEE----VEMSIDGFEKKLEGLFRRLI 272
> sce:YOR137C SIA1; Protein of unassigned function involved in
activation of the Pma1p plasma membrane H+-ATPase by glucose
Length=622
Score = 30.0 bits (66), Expect = 3.8, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 23/35 (65%), Gaps = 6/35 (17%)
Query 13 TRMAESFTVESTDAASSAFTLNSGPYRNIPIYVHL 47
TR++++ TVE+ ++L SGP+ N +YVHL
Sbjct 93 TRVSKNITVETL------YSLQSGPFYNSYLYVHL 121
Lambda K H
0.319 0.133 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4471152252
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40