bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3612_orf2
Length=184
Score E
Sequences producing significant alignments: (Bits) Value
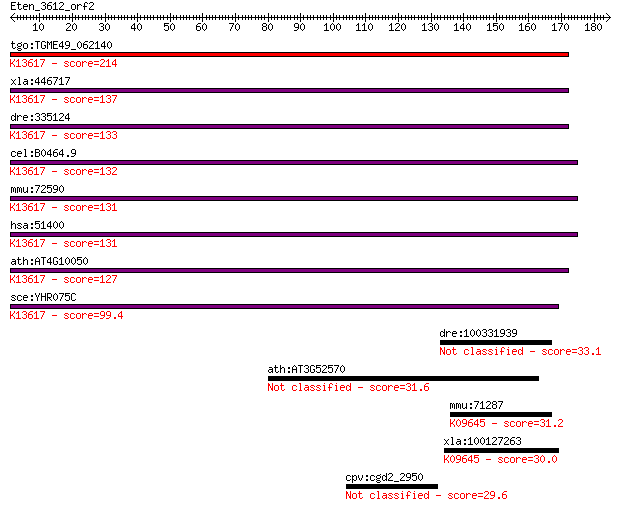
tgo:TGME49_062140 protein phosphatase methylesterase 1, putati... 214 2e-55
xla:446717 ppme1, MGC84506; protein phosphatase methylesterase... 137 3e-32
dre:335124 ppme1, fk72d03, wu:fk72d03, zgc:56239; protein phos... 133 4e-31
cel:B0464.9 hypothetical protein; K13617 protein phosphatase m... 132 5e-31
mmu:72590 Ppme1, 1110069N17Rik, 2700017M01Rik, PME-1, Pme1; pr... 131 1e-30
hsa:51400 PPME1, FLJ22226, PME-1; protein phosphatase methyles... 131 1e-30
ath:AT4G10050 hydrolase, alpha/beta fold family protein; K1361... 127 3e-29
sce:YHR075C PPE1; Ppe1p (EC:3.1.1.-); K13617 protein phosphata... 99.4 6e-21
dre:100331939 abhydrolase domain containing 9-like 33.1 0.49
ath:AT3G52570 ATP binding 31.6 1.7
mmu:71287 Cpvl, 4933436L16Rik, HVLP; carboxypeptidase, vitello... 31.2 2.3
xla:100127263 cpvl; carboxypeptidase, vitellogenic-like; K0964... 30.0 4.2
cpv:cgd2_2950 ring domain at very C-terminus of large protein 29.6 6.1
> tgo:TGME49_062140 protein phosphatase methylesterase 1, putative
; K13617 protein phosphatase methylesterase 1 [EC:3.1.1.-]
Length=452
Score = 214 bits (544), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 102/172 (59%), Positives = 130/172 (75%), Gaps = 1/172 (0%)
Query 1 LMVLDVVEGTALAALPQMAAFVSRFPSLFTSCKEAVNWSIFAGLLCNRSSAAISIPSQLV 60
L+V+DVVEGTA+AALP MA+F+ R P++FTS +EAV W+I +G L N SA +S+PSQLV
Sbjct 262 LVVVDVVEGTAMAALPHMASFIKRLPTVFTSPREAVCWAIRSGTLKNEESARVSLPSQLV 321
Query 61 KTTRGAL-PASHKGADKGPPDEEVWTWRVDVMATEPYWEGWFRGMSHAFLSSRCVKILIC 119
+T R + K G D+ VWTWR D+ T+P+WEGWF GMS FL +RC K+LIC
Sbjct 322 QTRRRDVRDILKKEISPGHEDDVVWTWRTDLAKTQPFWEGWFDGMSSLFLQARCTKVLIC 381
Query 120 SSSDRLDKELMIAHMQGKFQVQIVSGSGHVIEEDQPAELCRVVQTFITRYRL 171
+ +DRLD+ELMIAHMQGKFQVQ+V SGHV+EEDQP E+ V+ FI+RYRL
Sbjct 382 AGNDRLDRELMIAHMQGKFQVQLVPYSGHVVEEDQPQEVANVLLNFISRYRL 433
> xla:446717 ppme1, MGC84506; protein phosphatase methylesterase
1; K13617 protein phosphatase methylesterase 1 [EC:3.1.1.-]
Length=386
Score = 137 bits (344), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 72/198 (36%), Positives = 101/198 (51%), Gaps = 27/198 (13%)
Query 1 LMVLDVVEGTALAALPQMAAFVSRFPSLFTSCKEAVNWSIFAGLLCNRSSAAISIPSQLV 60
L V+DVVEGTA+ AL M F+ P F S + A+ WS+ +G + N SA +S+ Q+
Sbjct 175 LCVIDVVEGTAMDALNSMQNFLRSRPKTFKSLENAIEWSVKSGQIRNLESARVSMAGQIK 234
Query 61 KTTRGALPASHKGA---------------------------DKGPPDEEVWTWRVDVMAT 93
+ P K D E +TWR+++ T
Sbjct 235 QCEEATTPEGPKAIVEGIIEEEEEDEEENGGQSINKRKKEDDTDTKKERPYTWRIELSKT 294
Query 94 EPYWEGWFRGMSHAFLSSRCVKILICSSSDRLDKELMIAHMQGKFQVQIVSGSGHVIEED 153
E YWEGWFRG+S+ FLS K L+ + DRLDK+L I MQGKFQ+Q++ GH + ED
Sbjct 295 EKYWEGWFRGLSNLFLSCAIPKQLLLAGVDRLDKDLTIGQMQGKFQMQVLPQCGHAVHED 354
Query 154 QPAELCRVVQTFITRYRL 171
P ++ V TF+ R+R
Sbjct 355 APDKVAEAVATFLLRHRF 372
> dre:335124 ppme1, fk72d03, wu:fk72d03, zgc:56239; protein phosphatase
methylesterase 1 (EC:3.1.1.-); K13617 protein phosphatase
methylesterase 1 [EC:3.1.1.-]
Length=377
Score = 133 bits (334), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 72/200 (36%), Positives = 104/200 (52%), Gaps = 32/200 (16%)
Query 1 LMVLDVVEGTALAALPQMAAFVSRFPSLFTSCKEAVNWSIFAGLLCNRSSAAISIPSQLV 60
L V+DVVEGTA+ AL M F+ P F S + A+ WS+ +G + N SA +S+ Q+
Sbjct 173 LCVIDVVEGTAMDALNSMQNFLRSRPKTFKSVENAIEWSVKSGQIRNVESARVSMVGQVK 232
Query 61 KTTRGALPASHKGA-----------------------------DKGPPDEEVWTWRVDVM 91
K P S G D+ E ++TWR+++
Sbjct 233 KCEE---PLSSPGVSKSISEGIIEEEEEDEEGGESNHKRKKEDDQEIKKESLYTWRIELS 289
Query 92 ATEPYWEGWFRGMSHAFLSSRCVKILICSSSDRLDKELMIAHMQGKFQVQIVSGSGHVIE 151
TE YWEGWF+G+S FLS K+L+ + DRLDK+L I MQGKFQ+Q++ GH +
Sbjct 290 KTEKYWEGWFKGLSSLFLSCSVPKLLLLAGIDRLDKDLTIGQMQGKFQMQVLPQCGHAVH 349
Query 152 EDQPAELCRVVQTFITRYRL 171
ED P ++ + TF+ R+R
Sbjct 350 EDAPEKVADALATFMVRHRF 369
> cel:B0464.9 hypothetical protein; K13617 protein phosphatase
methylesterase 1 [EC:3.1.1.-]
Length=364
Score = 132 bits (333), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 66/174 (37%), Positives = 100/174 (57%), Gaps = 17/174 (9%)
Query 1 LMVLDVVEGTALAALPQMAAFVSRFPSLFTSCKEAVNWSIFAGLLCNRSSAAISIPSQLV 60
L+V+DVVEG+A+ AL M F+ PS F S ++A++W + +G N ++A +S+PSQ+
Sbjct 186 LIVIDVVEGSAMEALGGMVHFLHSRPSSFPSIEKAIHWCLSSGTARNPTAARVSMPSQIR 245
Query 61 KTTRGALPASHKGADKGPPDEEVWTWRVDVMATEPYWEGWFRGMSHAFLSSRCVKILICS 120
+ + E +TWR+D+ TE YW+GWF G+S FL K+L+ +
Sbjct 246 EVS-----------------EHEYTWRIDLTTTEQYWKGWFEGLSKEFLGCSVPKMLVLA 288
Query 121 SSDRLDKELMIAHMQGKFQVQIVSGSGHVIEEDQPAELCRVVQTFITRYRLHLP 174
DRLD++L I MQGKFQ ++ GH ++ED P L V F R+R+ P
Sbjct 289 GVDRLDRDLTIGQMQGKFQTCVLPKVGHCVQEDSPQNLADEVGRFACRHRIAQP 342
> mmu:72590 Ppme1, 1110069N17Rik, 2700017M01Rik, PME-1, Pme1;
protein phosphatase methylesterase 1 (EC:3.1.1.-); K13617 protein
phosphatase methylesterase 1 [EC:3.1.1.-]
Length=386
Score = 131 bits (330), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 70/199 (35%), Positives = 103/199 (51%), Gaps = 25/199 (12%)
Query 1 LMVLDVVEGTALAALPQMAAFVSRFPSLFTSCKEAVNWSIFAGLLCNRSSAAISIPSQLV 60
L ++DVVEGTA+ AL M F+ P F S + A+ WS+ +G + N SA +S+ Q+
Sbjct 177 LCMIDVVEGTAMDALNSMQNFLRGRPKTFKSLENAIEWSVKSGQIRNLESARVSMVGQVK 236
Query 61 KTTRGALPASHKGA-------------------------DKGPPDEEVWTWRVDVMATEP 95
+ P K D + +TWR+++ TE
Sbjct 237 QCEGITSPEGSKSIVEGIIEEEEEDEEGSESVNKRKKEDDMETKKDHPYTWRIELAKTEK 296
Query 96 YWEGWFRGMSHAFLSSRCVKILICSSSDRLDKELMIAHMQGKFQVQIVSGSGHVIEEDQP 155
YW+GWFRG+S+ FLS K+L+ + DRLDK+L I MQGKFQ+Q++ GH + ED P
Sbjct 297 YWDGWFRGLSNLFLSCPIPKLLLLAGVDRLDKDLTIGQMQGKFQMQVLPQCGHAVHEDAP 356
Query 156 AELCRVVQTFITRYRLHLP 174
++ V TF+ R+R P
Sbjct 357 DKVAEAVATFLIRHRFAEP 375
> hsa:51400 PPME1, FLJ22226, PME-1; protein phosphatase methylesterase
1 (EC:3.1.1.-); K13617 protein phosphatase methylesterase
1 [EC:3.1.1.-]
Length=386
Score = 131 bits (330), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 70/199 (35%), Positives = 103/199 (51%), Gaps = 25/199 (12%)
Query 1 LMVLDVVEGTALAALPQMAAFVSRFPSLFTSCKEAVNWSIFAGLLCNRSSAAISIPSQLV 60
L ++DVVEGTA+ AL M F+ P F S + A+ WS+ +G + N SA +S+ Q+
Sbjct 177 LCMIDVVEGTAMDALNSMQNFLRGRPKTFKSLENAIEWSVKSGQIRNLESARVSMVGQVK 236
Query 61 KTTRGALPASHKGA-------------------------DKGPPDEEVWTWRVDVMATEP 95
+ P K D + +TWR+++ TE
Sbjct 237 QCEGITSPEGSKSIVEGIIEEEEEDEEGSESISKRKKEDDMETKKDHPYTWRIELAKTEK 296
Query 96 YWEGWFRGMSHAFLSSRCVKILICSSSDRLDKELMIAHMQGKFQVQIVSGSGHVIEEDQP 155
YW+GWFRG+S+ FLS K+L+ + DRLDK+L I MQGKFQ+Q++ GH + ED P
Sbjct 297 YWDGWFRGLSNLFLSCPIPKLLLLAGVDRLDKDLTIGQMQGKFQMQVLPQCGHAVHEDAP 356
Query 156 AELCRVVQTFITRYRLHLP 174
++ V TF+ R+R P
Sbjct 357 DKVAEAVATFLIRHRFAEP 375
> ath:AT4G10050 hydrolase, alpha/beta fold family protein; K13617
protein phosphatase methylesterase 1 [EC:3.1.1.-]
Length=350
Score = 127 bits (318), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 66/171 (38%), Positives = 98/171 (57%), Gaps = 16/171 (9%)
Query 1 LMVLDVVEGTALAALPQMAAFVSRFPSLFTSCKEAVNWSIFAGLLCNRSSAAISIPSQLV 60
L+V+DVVEGTA+++L M +S F S ++A+ +S+ G L N SA +SIP+ L
Sbjct 177 LVVVDVVEGTAISSLIHMQKILSNRMQHFPSIEKAIEYSVRGGSLRNIDSARVSIPTTL- 235
Query 61 KTTRGALPASHKGADKGPPDEEVWTWRVDVMATEPYWEGWFRGMSHAFLSSRCVKILICS 120
K + + +R + TE YW+GW+ G+S FLSS K+L+ +
Sbjct 236 ---------------KYDDSKHCYVYRTRLEETEQYWKGWYDGLSEKFLSSPVPKLLLLA 280
Query 121 SSDRLDKELMIAHMQGKFQVQIVSGSGHVIEEDQPAELCRVVQTFITRYRL 171
+DRLD+ L I MQGKFQ+ +V +GH I+ED P E +V FI+R R+
Sbjct 281 GTDRLDRTLTIGQMQGKFQMIVVKHTGHAIQEDVPEEFANLVLNFISRNRI 331
> sce:YHR075C PPE1; Ppe1p (EC:3.1.1.-); K13617 protein phosphatase
methylesterase 1 [EC:3.1.1.-]
Length=400
Score = 99.4 bits (246), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 54/169 (31%), Positives = 88/169 (52%), Gaps = 19/169 (11%)
Query 1 LMVLDVVEGTALAALPQMAAFVSRFPSLFTSCKEAVNWSIFAGLLCNRSSAAISIPSQLV 60
+ +LD+VE A+ AL ++ F+ P++F S +AV+W + L RSSA I+IP+
Sbjct 229 ITMLDIVEEAAIMALNKVEHFLQNTPNVFESINDAVDWHVQHALSRLRSSAEIAIPALFA 288
Query 61 KTTRGALPASHKGADKGPPDEEVWTWRVDVMAT-EPYWEGWFRGMSHAFLSSRCVKILIC 119
G + R+ + T P+W+ WF +SH+F+ K+LI
Sbjct 289 PLKSGKVV------------------RITNLKTFSPFWDTWFTDLSHSFVGLPVSKLLIL 330
Query 120 SSSDRLDKELMIAHMQGKFQVQIVSGSGHVIEEDQPAELCRVVQTFITR 168
+ ++ LDKEL++ MQGK+Q+ + SGH I+ED P + + F R
Sbjct 331 AGNENLDKELIVGQMQGKYQLVVFQDSGHFIQEDSPIKTAITLIDFWKR 379
> dre:100331939 abhydrolase domain containing 9-like
Length=370
Score = 33.1 bits (74), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 11/34 (32%), Positives = 25/34 (73%), Gaps = 0/34 (0%)
Query 133 HMQGKFQVQIVSGSGHVIEEDQPAELCRVVQTFI 166
+++ F++ I+SG+ H +++DQP + +++ TFI
Sbjct 325 YIRNLFRLNIISGASHWLQQDQPDIVNKLIWTFI 358
> ath:AT3G52570 ATP binding
Length=335
Score = 31.6 bits (70), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 37/95 (38%), Gaps = 12/95 (12%)
Query 80 DEEVWTWRVD--VMATEPYWEGWFRGMSHAFLSSRCVKILICSSSDRLDKELM-----IA 132
D E WT+ +D V Y E + + + +I SDR D + IA
Sbjct 232 DSETWTFNLDGAVQMFNSYRETSYWSLLENPPKETEINFVIAEKSDRWDNDTTKRLETIA 291
Query 133 HM-----QGKFQVQIVSGSGHVIEEDQPAELCRVV 162
+ +GK ++ SGH + D P L +V
Sbjct 292 NQRQNVAEGKVATHLLRNSGHWVHTDNPKGLLEIV 326
> mmu:71287 Cpvl, 4933436L16Rik, HVLP; carboxypeptidase, vitellogenic-like
(EC:3.4.16.-); K09645 vitellogenic carboxypeptidase-like
protein [EC:3.4.16.-]
Length=478
Score = 31.2 bits (69), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 17/31 (54%), Gaps = 0/31 (0%)
Query 136 GKFQVQIVSGSGHVIEEDQPAELCRVVQTFI 166
GKF IV G GH++ DQP ++ FI
Sbjct 438 GKFHQVIVRGGGHILPYDQPMRSFDMINRFI 468
> xla:100127263 cpvl; carboxypeptidase, vitellogenic-like; K09645
vitellogenic carboxypeptidase-like protein [EC:3.4.16.-]
Length=481
Score = 30.0 bits (66), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 134 MQGKFQVQIVSGSGHVIEEDQPAELCRVVQTFITR 168
+ G+F IV G GH++ DQP ++ FI +
Sbjct 441 ITGEFSQVIVRGGGHILPYDQPERTYSMIDRFIRK 475
> cpv:cgd2_2950 ring domain at very C-terminus of large protein
Length=2162
Score = 29.6 bits (65), Expect = 6.1, Method: Composition-based stats.
Identities = 12/28 (42%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 104 MSHAFLSSRCVKILICSSSDRLDKELMI 131
+ H F+S +K+L CS LDKEL +
Sbjct 1135 VKHLFISEESIKLLSCSKEISLDKELFL 1162
Lambda K H
0.322 0.134 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5041515336
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40