bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3634_orf1
Length=132
Score E
Sequences producing significant alignments: (Bits) Value
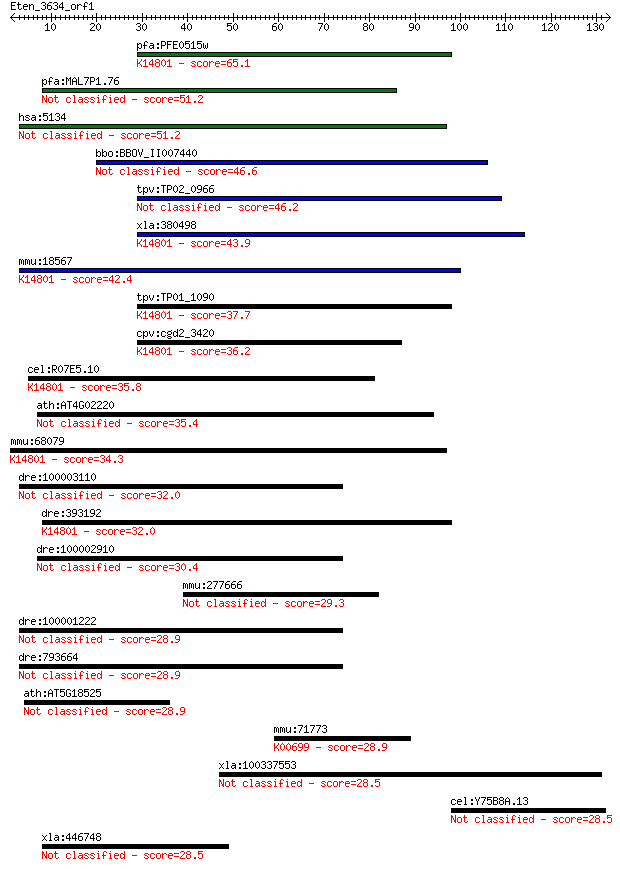
pfa:PFE0515w conserved Plasmodium protein, unknown function; K... 65.1 5e-11
pfa:MAL7P1.76 conserved Plasmodium protein, unknown function 51.2 7e-07
hsa:5134 PDCD2, MGC12347, RP8, ZMYND7; programmed cell death 2 51.2
bbo:BBOV_II007440 18.m06617; ubiquinone biosynthesis O-methylt... 46.6 2e-05
tpv:TP02_0966 hypothetical protein 46.2 2e-05
xla:380498 pdcd2, MGC53229; programmed cell death 2; K14801 pr... 43.9 1e-04
mmu:18567 Pdcd2, AI528543, Pcd2, RP-8; programmed cell death 2... 42.4 4e-04
tpv:TP01_1090 hypothetical protein; K14801 pre-rRNA-processing... 37.7 0.010
cpv:cgd2_3420 programmed cell death 2 ; K14801 pre-rRNA-proces... 36.2 0.029
cel:R07E5.10 pdcd-2; PDCD (mammalian ProgrammeD Cell Death pro... 35.8 0.035
ath:AT4G02220 zinc finger (MYND type) family protein / program... 35.4 0.048
mmu:68079 Pdcd2l, 6030457N17Rik, MGC144595; programmed cell de... 34.3 0.090
dre:100003110 novel protein containing lectin C-type domains 32.0 0.54
dre:393192 MGC56288; zgc:56288; K14801 pre-rRNA-processing pro... 32.0 0.57
dre:100002910 novel protein containing lectin C-type domains 30.4 1.4
mmu:277666 Gm13078, OTTMUSG00000010322; predicted gene 13078 29.3
dre:100001222 mannose receptor C type 1-like 28.9 3.8
dre:793664 mannose receptor C type 1-like 28.9 4.4
ath:AT5G18525 ATP binding / protein kinase/ protein serine/thr... 28.9 4.6
mmu:71773 Ugt2b1, 1300012D20Rik; UDP glucuronosyltransferase 2... 28.9 4.7
xla:100337553 sall4b, Xsal-3; zinc finger transcription factor... 28.5 5.2
cel:Y75B8A.13 hypothetical protein 28.5 5.6
xla:446748 wbp4, MGC78936; WW domain binding protein 4 (formin... 28.5 6.1
> pfa:PFE0515w conserved Plasmodium protein, unknown function;
K14801 pre-rRNA-processing protein TSR4
Length=439
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 27/69 (39%), Positives = 35/69 (50%), Gaps = 0/69 (0%)
Query 29 SKFGGNPVWIDGKIPDGFPFNCSLCGVVLDFVCQVATPYGANKRCLYFFCCHAQATCGKR 88
SKFGG P+WI K P CS+C L F+ Q++T Y R LY FCC + C
Sbjct 20 SKFGGEPIWIHDKEPTNLNLKCSICKKDLTFLFQLSTSYDEYIRVLYLFCCMNSSKCNMN 79
Query 89 PEGWKVLRG 97
W ++G
Sbjct 80 KNNWVCIKG 88
> pfa:MAL7P1.76 conserved Plasmodium protein, unknown function
Length=832
Score = 51.2 bits (121), Expect = 7e-07, Method: Composition-based stats.
Identities = 32/92 (34%), Positives = 45/92 (48%), Gaps = 14/92 (15%)
Query 8 VLIGFFGE------SVPDGDQTAAEELSKFGGNPVWIDG-KIPDGFPFNCSLCGVVLDFV 60
VL+G+ E + D + + +SK GG P W+D +PD FNCS+C ++ F+
Sbjct 4 VLLGYLDEKKKRKNELRDKHKINKKFVSKIGGKPFWLDRINLPDEKEFNCSVCNNMMIFL 63
Query 61 CQVATPY----GANKRCLY-FFCCHA--QATC 85
Q+ P RCLY F C H QA C
Sbjct 64 LQIYAPLDELGNCFHRCLYVFICIHCGDQAKC 95
> hsa:5134 PDCD2, MGC12347, RP8, ZMYND7; programmed cell death
2
Length=221
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 35/99 (35%), Positives = 44/99 (44%), Gaps = 10/99 (10%)
Query 3 ASETPVLIGFFGESVPDGDQTAAEELSKFGGNPVWIDGK-IPDGFPFNCSLCGVVLDFVC 61
A PV +GF ES P + + SK GG P W+ +P C LCG L F+
Sbjct 4 AGARPVELGF-AESAPAWRLRSEQFPSKVGGRPAWLGAAGLPGPQALACELCGRPLSFLL 62
Query 62 QVATPY----GANKRCLYFFCCHAQATCGKRPEGWKVLR 96
QV P A RC++ FCC Q C G +V R
Sbjct 63 QVYAPLPGRPDAFHRCIFLFCCREQPCCA----GLRVFR 97
> bbo:BBOV_II007440 18.m06617; ubiquinone biosynthesis O-methyltransferase
family protein (EC:2.1.1.-)
Length=556
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 30/89 (33%), Positives = 45/89 (50%), Gaps = 9/89 (10%)
Query 20 GDQTAAEELSKFGGNPVWIDGKIPDGFPFNCSLCGVVLDFVCQVATPYGAN-KRCLYFFC 78
G+ A S+FGGNPV + + P C++C L F+ Q+ATPY + R L +
Sbjct 306 GEAFEAFRHSRFGGNPV-LPTSVQVATPPRCAVCQRELLFLMQLATPYKHDVDRVLVIYF 364
Query 79 C--HAQATCGKRPEGWKVLRGVVEPSAPA 105
C H++A+ EGW + R + S P
Sbjct 365 CPEHSEAS-----EGWSIYRYTMSSSVPG 388
> tpv:TP02_0966 hypothetical protein
Length=332
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/82 (35%), Positives = 43/82 (52%), Gaps = 10/82 (12%)
Query 29 SKFGGNPVW-IDGKIPDGFPFNCSLCGVVLDFVCQVATPYGANK-RCLYFFCCHAQATCG 86
SK G+PV+ ++ + D C +C + + F+ Q++ P AN+ R LY F C AT
Sbjct 22 SKLLGHPVYPVNLNVDDP---KCEICNLNMSFLMQLSAPTTANRNRVLYIFYCLNDATKD 78
Query 87 KRPEGWKVLRGVVEPSAPAQAE 108
K GWK+LR E P+ E
Sbjct 79 K---GWKLLRYSAE--KPSTTE 95
> xla:380498 pdcd2, MGC53229; programmed cell death 2; K14801
pre-rRNA-processing protein TSR4
Length=361
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 44/89 (49%), Gaps = 9/89 (10%)
Query 29 SKFGGNPVWI-DGKIPDGFPFNCSLCGVVLDFVCQVATP-YGANKRCLYFFCCHAQATCG 86
SK GG P W+ + +P+ C LCG L F+ QV P G+ R ++ FCC +C
Sbjct 28 SKVGGRPAWLGEVGVPEPAALQCGLCGKPLAFLLQVYAPCAGSFHRTIFIFCCR-NGSCH 86
Query 87 KRPEG--WKVLRGVVEPSAPAQAEAEAYG 113
+ E +KV R + P + + +YG
Sbjct 87 RESEAQCFKVYRNQL----PRKNDTYSYG 111
> mmu:18567 Pdcd2, AI528543, Pcd2, RP-8; programmed cell death
2; K14801 pre-rRNA-processing protein TSR4
Length=343
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 44/102 (43%), Gaps = 10/102 (9%)
Query 3 ASETPVLIGFFGESVPDGDQTAAEELSKFGGNPVWID-GKIPDGFPFNCSLCGVVLDFVC 61
A+ PV +GF E P + + SK GG P W+ ++P C+ CG L F+
Sbjct 4 AAPGPVELGF-AEEAPAWRLRSEQFPSKVGGRPAWLGLAELPGPGALACARCGRPLAFLL 62
Query 62 QVATPY----GANKRCLYFFCCHAQATCGKRPEGWKVLRGVV 99
QV P A R L+ FCC C G +V R +
Sbjct 63 QVYAPLPGRDDAFHRSLFLFCCREPPCCA----GLRVFRNQL 100
> tpv:TP01_1090 hypothetical protein; K14801 pre-rRNA-processing
protein TSR4
Length=372
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 31/76 (40%), Gaps = 13/76 (17%)
Query 29 SKFGGNPVWIDG-KIPDGFPFNCSLCGVVLDFVCQVATP------YGANKRCLYFFCCHA 81
SKFGG P W+D +P C C ++ F Q+ P + R +Y F C
Sbjct 30 SKFGGLPAWLDPVHLPKESELKCEKCQSIMTFFLQIYAPDDLCEENDSFHRTVYLFVCQP 89
Query 82 QATCGKRPEGWKVLRG 97
CG + WK R
Sbjct 90 ---CGNQ---WKAFRS 99
> cpv:cgd2_3420 programmed cell death 2 ; K14801 pre-rRNA-processing
protein TSR4
Length=329
Score = 36.2 bits (82), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 30/63 (47%), Gaps = 6/63 (9%)
Query 29 SKFGGNPVWIDGK-IPDGFPFNCSLCGVVLDFVCQVATPYGANK----RCLYFFCCHAQA 83
SKFGG P W++ + +P C+ CG + F+ QV P + R ++ F C
Sbjct 25 SKFGGKPAWLNPQNLPQYRDLQCNSCGTRMRFLLQVYAPQDDREDLFHRSIFLFIC-TNC 83
Query 84 TCG 86
TC
Sbjct 84 TCS 86
> cel:R07E5.10 pdcd-2; PDCD (mammalian ProgrammeD Cell Death protein)
homolog family member (pdcd-2); K14801 pre-rRNA-processing
protein TSR4
Length=386
Score = 35.8 bits (81), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 38/86 (44%), Gaps = 10/86 (11%)
Query 5 ETPVLIGFFGESVPDGDQTAAEE---LSKFGGNPVWIDGK-IPDGFPFNCSLCGVVLDFV 60
+ PV +GF + PD + L K GG P W++ K +P C++C L F+
Sbjct 7 DEPVNLGFGVQFEPDDLYRLRSQFLPLGKIGGKPSWLNPKNLPKSADLLCNVCEKPLCFL 66
Query 61 CQVATPYGAN------KRCLYFFCCH 80
QV+ G N R L+ F C
Sbjct 67 MQVSANGGINDPPHAFHRSLFLFVCR 92
> ath:AT4G02220 zinc finger (MYND type) family protein / programmed
cell death 2 C-terminal domain-containing protein
Length=418
Score = 35.4 bits (80), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 28/98 (28%), Positives = 42/98 (42%), Gaps = 16/98 (16%)
Query 7 PVLIGFFGESVPDGDQTAAEELSKF-----GGNPVWIDG-KIPDGFPFNCSLCGVVLDFV 60
PV++GF + + A L + GG P W+D +P G C LC + FV
Sbjct 51 PVMLGFV-----ESPKFAWSNLRQLFPNLAGGVPAWLDPVNLPSGKSILCDLCEEPMQFV 105
Query 61 CQVATPY----GANKRCLYFFCCHAQATCGK-RPEGWK 93
Q+ P A R L+ F C + + + + E WK
Sbjct 106 LQLYAPLTDKESAFHRTLFLFMCPSMSCLLRDQHEQWK 143
> mmu:68079 Pdcd2l, 6030457N17Rik, MGC144595; programmed cell
death 2-like; K14801 pre-rRNA-processing protein TSR4
Length=364
Score = 34.3 bits (77), Expect = 0.090, Method: Compositional matrix adjust.
Identities = 35/108 (32%), Positives = 43/108 (39%), Gaps = 23/108 (21%)
Query 1 IMASETPVLIGFFGESV---PDGDQTAAEELSKFGGNPVWIDGKIPDGFPF------NCS 51
+ A PVL+G +V P G +A SK GG +PD P C
Sbjct 1 MAAVRKPVLLGLRDTAVKGCPKG--PSAWTSSKLGG--------VPDALPAVTTPGPQCG 50
Query 52 LCGVVLDFVCQVATPYGAN--KRCLYFFCCHAQATCGK-RPEGWKVLR 96
C L V QV P + R LY F C A+ CG + WKV R
Sbjct 51 RCAQPLTLVVQVYCPLDGSPFHRLLYVFAC-ARPGCGNSQTRSWKVFR 97
> dre:100003110 novel protein containing lectin C-type domains
Length=362
Score = 32.0 bits (71), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 27/71 (38%), Gaps = 5/71 (7%)
Query 3 ASETPVLIGFFGESVPDGDQTAAEELSKFGGNPVWIDGKIPDGFPFNCSLCGVVLDFVCQ 62
+S P L +G PDG A F N W DGK D + F C L FV Q
Sbjct 82 SSGDPALFLHWGNEQPDGADECA-----FMRNGQWEDGKCNDSWIFVCYNMSRGLVFVNQ 136
Query 63 VATPYGANKRC 73
T A C
Sbjct 137 AMTWRAAQSYC 147
> dre:393192 MGC56288; zgc:56288; K14801 pre-rRNA-processing protein
TSR4
Length=357
Score = 32.0 bits (71), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 30/92 (32%), Positives = 41/92 (44%), Gaps = 6/92 (6%)
Query 8 VLIGFFGESVPDGDQTAAEELSKFGGNPVWIDGKIPDGFPFNCSLCGVVLDFVCQVATPY 67
VLIG ++ D + A+ +K G P + I +P +CSLC L V QV P
Sbjct 9 VLIGICDGAI-DKKKNASYCTNKIGDRPDLL-PIITLQYP-SCSLCQRGLSHVVQVYCPL 65
Query 68 GAN--KRCLYFFCCHAQATCGKRPEGWKVLRG 97
A+ R + F C C + E W VLR
Sbjct 66 AASPYHRTINVFAC-TSPQCYGKSESWIVLRS 96
> dre:100002910 novel protein containing lectin C-type domains
Length=361
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 25/67 (37%), Gaps = 5/67 (7%)
Query 7 PVLIGFFGESVPDGDQTAAEELSKFGGNPVWIDGKIPDGFPFNCSLCGVVLDFVCQVATP 66
P L +G+ PDG A F N W DGK D + F C L FV Q
Sbjct 84 PALFLNWGDKQPDGKDECA-----FMKNGEWHDGKCSDTWIFVCYNMSRGLVFVNQKMNW 138
Query 67 YGANKRC 73
A C
Sbjct 139 RAAQSHC 145
> mmu:277666 Gm13078, OTTMUSG00000010322; predicted gene 13078
Length=474
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 19/45 (42%), Positives = 22/45 (48%), Gaps = 2/45 (4%)
Query 39 DGKIPDGFPFNCSLCGVVLDFVCQVATPYGANKRCLYF--FCCHA 81
DG IPD F CS L V Q Y +KRCLY FC ++
Sbjct 425 DGVIPDRFLQLCSELIDTLKVVRQPKQVYFVSKRCLYCRNFCIYS 469
> dre:100001222 mannose receptor C type 1-like
Length=360
Score = 28.9 bits (63), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 28/71 (39%), Gaps = 5/71 (7%)
Query 3 ASETPVLIGFFGESVPDGDQTAAEELSKFGGNPVWIDGKIPDGFPFNCSLCGVVLDFVCQ 62
+S P L +G + PDG + L F N W DGK D F C L +V Q
Sbjct 80 SSGDPALFLNWGYNQPDG-----KNLCAFMRNGEWYDGKCSDSCIFVCYNMSRGLVYVNQ 134
Query 63 VATPYGANKRC 73
+ A C
Sbjct 135 MMNWKSAQSYC 145
> dre:793664 mannose receptor C type 1-like
Length=322
Score = 28.9 bits (63), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 28/71 (39%), Gaps = 5/71 (7%)
Query 3 ASETPVLIGFFGESVPDGDQTAAEELSKFGGNPVWIDGKIPDGFPFNCSLCGVVLDFVCQ 62
+S P L +G + PDG + L F N W DGK D F C L +V Q
Sbjct 80 SSGDPALFLNWGYNQPDG-----KNLCAFMKNGEWYDGKCSDSCIFVCYNMSRGLVYVNQ 134
Query 63 VATPYGANKRC 73
+ A C
Sbjct 135 MMNWKSAQSYC 145
> ath:AT5G18525 ATP binding / protein kinase/ protein serine/threonine
kinase/ protein tyrosine kinase
Length=1639
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 14/32 (43%), Positives = 18/32 (56%), Gaps = 0/32 (0%)
Query 4 SETPVLIGFFGESVPDGDQTAAEELSKFGGNP 35
S+T LI F ES D DQ +++ SK NP
Sbjct 1365 SQTGKLISLFSESPSDQDQASSDPSSKNNSNP 1396
> mmu:71773 Ugt2b1, 1300012D20Rik; UDP glucuronosyltransferase
2 family, polypeptide B1 (EC:2.4.1.17); K00699 glucuronosyltransferase
[EC:2.4.1.17]
Length=529
Score = 28.9 bits (63), Expect = 4.7, Method: Composition-based stats.
Identities = 14/30 (46%), Positives = 16/30 (53%), Gaps = 2/30 (6%)
Query 59 FVCQVATPYGANKRCLYFFCCHAQATCGKR 88
VC VA + K CL FCCH A GK+
Sbjct 499 LVCVVAVVFIIAKCCL--FCCHKTANMGKK 526
> xla:100337553 sall4b, Xsal-3; zinc finger transcription factor
SALL4 b
Length=1061
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 26/101 (25%), Positives = 37/101 (36%), Gaps = 17/101 (16%)
Query 47 PFNCSLCGVVLDFVCQVATPYGANKRCLYFFCCHAQATCGKRPEGWKVLRGVVE------ 100
PF C +CG + T YG ++ H+ C K+ VL+ +
Sbjct 564 PFKCKICGRAFSTKSNLKTHYGVHRANTPLKLQHSCPICQKKFTNAVVLQQHIRMHMGGK 623
Query 101 -PSAPAQAEA---------EAYGAAHTSFKREN-DDLAAAD 130
P+ P EA E G + SF EN DD+ D
Sbjct 624 IPNTPVSEEASDDIDSMMDEKNGELNNSFTDENLDDIDMED 664
> cel:Y75B8A.13 hypothetical protein
Length=1420
Score = 28.5 bits (62), Expect = 5.6, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 98 VVEPSAPAQAEAEAYGAAHTSFKRENDDLAAADP 131
V P+APAQ E+ A F E+D+ A DP
Sbjct 1381 VTTPAAPAQESTESSAAQKREFLPEHDEDEAQDP 1414
> xla:446748 wbp4, MGC78936; WW domain binding protein 4 (formin
binding protein 21)
Length=374
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 8 VLIGFFGESVPDGDQTAAEELSKFGGNPVWIDGKIPDGFPF 48
L G P+G Q +EE +K G VW++G +GF +
Sbjct 136 TLTGESQWEEPEGFQDKSEESNKGGSGSVWVEGLSEEGFTY 176
Lambda K H
0.320 0.137 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2099897216
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40