bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3647_orf2
Length=197
Score E
Sequences producing significant alignments: (Bits) Value
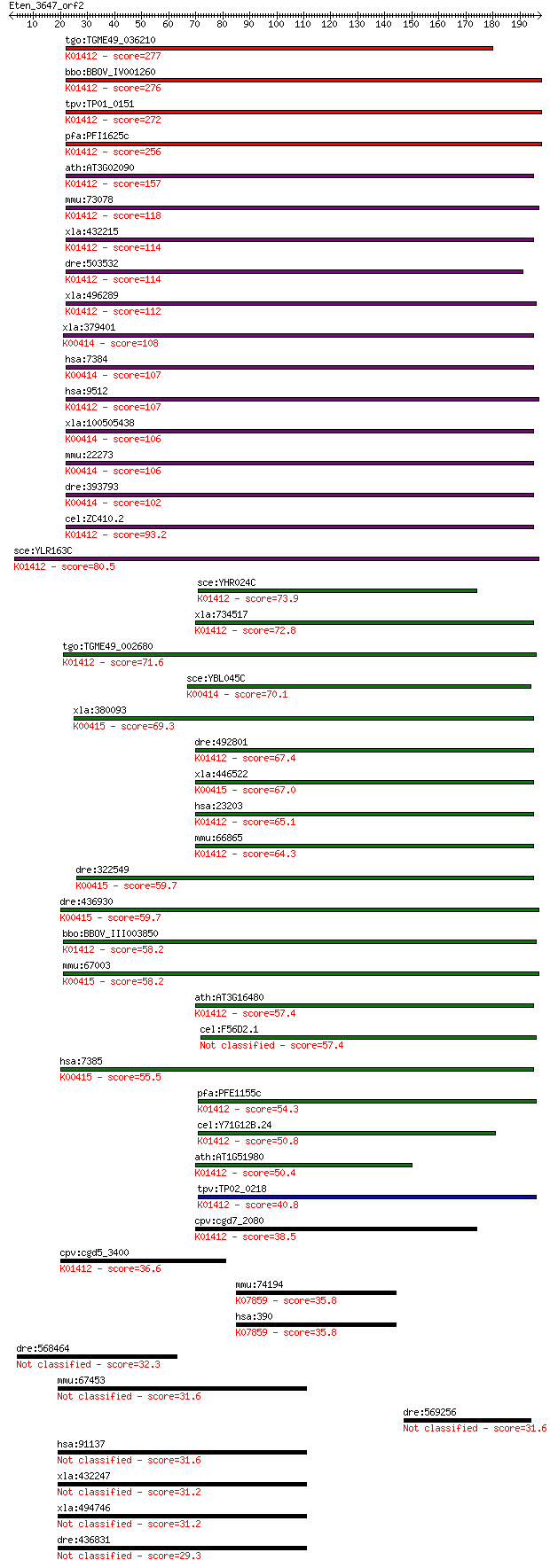
tgo:TGME49_036210 mitochondrial-processing peptidase beta subu... 277 1e-74
bbo:BBOV_IV001260 21.m02910; mitochondrial processing peptidas... 276 3e-74
tpv:TP01_0151 biquinol-cytochrome C reductase complex core pro... 272 6e-73
pfa:PFI1625c organelle processing peptidase, putative; K01412 ... 256 4e-68
ath:AT3G02090 MPPBETA; mitochondrial processing peptidase beta... 157 2e-38
mmu:73078 Pmpcb, 3110004O18Rik, MPP11, MPPB, MPPP52; peptidase... 118 1e-26
xla:432215 pmpcb, MGC78954; peptidase (mitochondrial processin... 114 2e-25
dre:503532 pmpcb, zgc:110738; peptidase (mitochondrial process... 114 3e-25
xla:496289 hypothetical LOC496289; K01412 mitochondrial proces... 112 6e-25
xla:379401 uqcrc1, MGC53748; ubiquinol-cytochrome c reductase ... 108 9e-24
hsa:7384 UQCRC1, D3S3191, QCR1, UQCR1; ubiquinol-cytochrome c ... 107 2e-23
hsa:9512 PMPCB, Beta-MPP, MPP11, MPPB, MPPP52, P-52; peptidase... 107 2e-23
xla:100505438 hypothetical protein LOC100505438; K00414 ubiqui... 106 4e-23
mmu:22273 Uqcrc1, 1110032G10Rik, MGC97899; ubiquinol-cytochrom... 106 4e-23
dre:393793 uqcrc1, MGC73404, zgc:73404, zgc:85750; ubiquinol-c... 102 1e-21
cel:ZC410.2 mppb-1; Mitochondrial Processing Peptidase Beta fa... 93.2 6e-19
sce:YLR163C MAS1, MIF1; Mas1p (EC:3.4.24.64); K01412 mitochond... 80.5 3e-15
sce:YHR024C MAS2, MIF2; Mas2p (EC:3.4.24.64); K01412 mitochond... 73.9 3e-13
xla:734517 pmpca, MGC114896; peptidase (mitochondrial processi... 72.8 8e-13
tgo:TGME49_002680 mitochondrial-processing peptidase alpha sub... 71.6 1e-12
sce:YBL045C COR1, QCR1; Cor1p; K00414 ubiquinol-cytochrome c r... 70.1 4e-12
xla:380093 uqcrc2, MGC53943; Ubiquinol-cytochrome C reductase ... 69.3 9e-12
dre:492801 pmpca, wu:fi19e06, wu:fj83d11, zgc:101647; peptidas... 67.4 3e-11
xla:446522 uqcrc2, MGC130698, MGC80228; ubiquinol-cytochrome c... 67.0 4e-11
hsa:23203 PMPCA, Alpha-MPP, FLJ26258, INPP5E, KIAA0123, MGC104... 65.1 1e-10
mmu:66865 Pmpca, 1200002L24Rik, 4933435E07Rik, Alpha-MPP, P-55... 64.3 3e-10
dre:322549 uqcrc2b, fi06a11, uqcrc2, wu:fb64g04, wu:fi06a11; u... 59.7 6e-09
dre:436930 uqcrc2a, fb33d01, wu:fb33d01, wu:fc43c05, zgc:92453... 59.7 7e-09
bbo:BBOV_III003850 17.m07356; mitochondrial processing peptida... 58.2 2e-08
mmu:67003 Uqcrc2, 1500004O06Rik; ubiquinol cytochrome c reduct... 58.2 2e-08
ath:AT3G16480 MPPalpha (mitochondrial processing peptidase alp... 57.4 3e-08
cel:F56D2.1 ucr-1; Ubiquinol-Cytochrome c oxidoReductase compl... 57.4 3e-08
hsa:7385 UQCRC2, QCR2, UQCR2; ubiquinol-cytochrome c reductase... 55.5 1e-07
pfa:PFE1155c mitochondrial processing peptidase alpha subunit,... 54.3 2e-07
cel:Y71G12B.24 mppa-1; Mitochondrial Processing Peptidase Alph... 50.8 3e-06
ath:AT1G51980 mitochondrial processing peptidase alpha subunit... 50.4 4e-06
tpv:TP02_0218 ubiquinol-cytochrome C reductase complex core pr... 40.8 0.003
cpv:cgd7_2080 mitochondrial processing peptidase, insulinase l... 38.5 0.014
cpv:cgd5_3400 mitochondrial processing peptidase beta subunit ... 36.6 0.059
mmu:74194 Rnd3, 2610017M01Rik, AI661404, Arhe, Rhoe; Rho famil... 35.8 0.100
hsa:390 RND3, ARHE, Rho8, RhoE, memB; Rho family GTPase 3; K07... 35.8 0.100
dre:568464 zfpm2b, FOG-B, fog2b; zinc finger protein, multityp... 32.3 1.1
mmu:67453 Slc25a46, 1200007B05Rik, AI325987; solute carrier fa... 31.6 1.9
dre:569256 MGC153498; zgc:153498 31.6 2.0
hsa:91137 SLC25A46; solute carrier family 25, member 46 31.6
xla:432247 slc25a46-a, MGC81360; solute carrier family 25, mem... 31.2 2.1
xla:494746 slc25a46-b, slc25a46; solute carrier family 25, mem... 31.2 2.6
dre:436831 slc25a46, zgc:92767; solute carrier family 25, memb... 29.3 8.5
> tgo:TGME49_036210 mitochondrial-processing peptidase beta subunit,
putative (EC:3.4.24.64); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=524
Score = 277 bits (709), Expect = 1e-74, Method: Compositional matrix adjust.
Identities = 124/158 (78%), Positives = 142/158 (89%), Gaps = 0/158 (0%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTGL 81
D V FMLMQAI+GSYRKH+EG+VPGK+SAN TVRN+ KM GCA+MFSAFNTCY DTGL
Sbjct 323 DAVTFMLMQAIVGSYRKHDEGIVPGKVSANATVRNVCNKMTVGCADMFSAFNTCYSDTGL 382
Query 82 FGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSVAEDL 141
FGFYAQCDEVA+EHCV+++M GIT+LSY+VTDEEVERAKAQL TQLLGHLDSTT+VAED+
Sbjct 383 FGFYAQCDEVALEHCVMEIMFGITSLSYAVTDEEVERAKAQLKTQLLGHLDSTTAVAEDI 442
Query 142 GRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHD 179
GRQ+L YGR +P AEFL RL IDA+E+KRVAWKYLHD
Sbjct 443 GRQMLAYGRRMPLAEFLKRLEVIDAEEVKRVAWKYLHD 480
> bbo:BBOV_IV001260 21.m02910; mitochondrial processing peptidase
beta subunit; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=514
Score = 276 bits (706), Expect = 3e-74, Method: Compositional matrix adjust.
Identities = 125/176 (71%), Positives = 149/176 (84%), Gaps = 0/176 (0%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTGL 81
D V FMLMQ+IIGSY+K++EG+VPGK+S N TV IA +M GCAE FSAFNTCY+DTGL
Sbjct 328 DSVCFMLMQSIIGSYKKNQEGIVPGKVSGNKTVHAIANRMTVGCAEAFSAFNTCYKDTGL 387
Query 82 FGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSVAEDL 141
FGFYAQCDEVAV+HCV +LM G+T++SYS+TDEEVERAK QL Q L DST++VAE++
Sbjct 388 FGFYAQCDEVAVDHCVGELMFGVTSMSYSITDEEVERAKRQLMLQFLSMNDSTSTVAEEV 447
Query 142 GRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMPQLI 197
RQI+VYGR +P EFL+RL IDA+E+KRVAWKYLHD EVAV+AMGPLHGMP LI
Sbjct 448 ARQIIVYGRRMPVTEFLLRLEQIDAEEVKRVAWKYLHDHEVAVTAMGPLHGMPSLI 503
> tpv:TP01_0151 biquinol-cytochrome C reductase complex core protein
I (EC:1.10.2.2); K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=518
Score = 272 bits (695), Expect = 6e-73, Method: Compositional matrix adjust.
Identities = 125/176 (71%), Positives = 147/176 (83%), Gaps = 0/176 (0%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTGL 81
D VAFMLMQ+IIG+Y K EG+VPGK+S N T+ +A +M GCAE FSAFNTCY+DTGL
Sbjct 332 DSVAFMLMQSIIGTYNKSNEGVVPGKVSGNKTIHAVANRMTVGCAEFFSAFNTCYKDTGL 391
Query 82 FGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSVAEDL 141
FGFYA+ DEVAV+HCV +L+ GIT+LSYSVTDEEVERAK QL Q L +ST+SVAE++
Sbjct 392 FGFYAKADEVAVDHCVGELLFGITSLSYSVTDEEVERAKRQLMLQFLSMTESTSSVAEEV 451
Query 142 GRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMPQLI 197
RQILVYGR +P AEFL+RL IDA+E+KRVAWKYLHD EVAVSAMGPLHGMP L+
Sbjct 452 ARQILVYGRRMPVAEFLLRLEKIDAEEVKRVAWKYLHDSEVAVSAMGPLHGMPSLV 507
> pfa:PFI1625c organelle processing peptidase, putative; K01412
mitochondrial processing peptidase [EC:3.4.24.64]
Length=484
Score = 256 bits (653), Expect = 4e-68, Method: Compositional matrix adjust.
Identities = 111/176 (63%), Positives = 146/176 (82%), Gaps = 0/176 (0%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTGL 81
D + FMLMQ IIG+Y+K+EEG++PGKLSAN TV NI KM GCA+ F++FNTCY +TGL
Sbjct 298 DSITFMLMQCIIGTYKKNEEGILPGKLSANRTVNNICNKMTVGCADYFTSFNTCYNNTGL 357
Query 82 FGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSVAEDL 141
FGFY QCDE+AVEH + +LM G+T+LSYS+TDEEVE AK L TQL+ +S++++AE++
Sbjct 358 FGFYVQCDEIAVEHALGELMFGVTSLSYSITDEEVELAKIHLKTQLISMFESSSTLAEEV 417
Query 142 GRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMPQLI 197
RQ+LVYGR + AEF++RLN ID +E+KRVAWKYLHD+++AV+A+G LHGMPQ I
Sbjct 418 SRQLLVYGRKISLAEFILRLNEIDTEEVKRVAWKYLHDRDIAVAAIGALHGMPQYI 473
> ath:AT3G02090 MPPBETA; mitochondrial processing peptidase beta
subunit, putative; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=531
Score = 157 bits (397), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 81/177 (45%), Positives = 116/177 (65%), Gaps = 14/177 (7%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTGL 81
D VA M+MQ ++GS+ K+ G GK + + +A AE AFNT Y+DTGL
Sbjct 351 DSVALMVMQTMLGSWNKNVGG---GKHVGSDLTQRVAI---NEIAESIMAFNTNYKDTGL 404
Query 82 FGFYAQCDEVAVEHCVLDL----MHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSV 137
FG YA VA C+ DL M+ +T L+Y V+D +V RA+ QL + LL H+D T+ +
Sbjct 405 FGVYA----VAKADCLDDLSYAIMYEVTKLAYRVSDADVTRARNQLKSSLLLHMDGTSPI 460
Query 138 AEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMP 194
AED+GRQ+L YGR +PTAE R++A+DA +KRVA KY++DK++A+SA+GP+ +P
Sbjct 461 AEDIGRQLLTYGRRIPTAELFARIDAVDASTVKRVANKYIYDKDIAISAIGPIQDLP 517
> mmu:73078 Pmpcb, 3110004O18Rik, MPP11, MPPB, MPPP52; peptidase
(mitochondrial processing) beta (EC:3.4.24.64); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=489
Score = 118 bits (295), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 62/180 (34%), Positives = 99/180 (55%), Gaps = 16/180 (8%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKM-QTGC----AEMFSAFNTCY 76
D + M+ +IG++ + G + N+++K+ Q C F +FNT Y
Sbjct 309 DTICLMVANTLIGNWDRSFGGGM-----------NLSSKLAQLTCHGNLCHSFQSFNTSY 357
Query 77 RDTGLFGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTS 136
DTGL+G Y C++ V + + + L VT+ EV RAK L T +L LD +T
Sbjct 358 TDTGLWGLYMVCEQATVADMLHVVQNEWKRLCTDVTESEVARAKNLLKTNMLLQLDGSTP 417
Query 137 VAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMPQL 196
+ ED+GRQ+L Y R +P E R++A+DA+ ++RV KY+HDK A++A+GP+ +P
Sbjct 418 ICEDIGRQMLCYNRRIPIPELEARIDAVDAETVRRVCTKYIHDKSPAIAALGPIERLPDF 477
> xla:432215 pmpcb, MGC78954; peptidase (mitochondrial processing)
beta (EC:3.4.24.64); K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=479
Score = 114 bits (286), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 67/182 (36%), Positives = 100/182 (54%), Gaps = 24/182 (13%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKM-QTGC----AEMFSAFNTCY 76
D + M+ +IG++ + G V N+++K+ Q C F +FNTCY
Sbjct 299 DTIPLMVANTLIGNWDRSFGGGV-----------NLSSKLAQLTCHGNLCHSFQSFNTCY 347
Query 77 RDTGLFGFYAQCDEVAVEHCVLDLMHGIT----ALSYSVTDEEVERAKAQLTTQLLGHLD 132
DTGL+G Y C+ VE D+MH + L SVT+ EV RAK L T +L LD
Sbjct 348 TDTGLWGLYMVCEPNTVE----DMMHFVQREWIRLCTSVTENEVARAKNLLKTNMLLQLD 403
Query 133 STTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHG 192
+T + ED+GRQ+L Y R +P E R++ I A+ I+ V KY+++K AV+A+GP+
Sbjct 404 GSTPICEDIGRQMLCYNRRIPLPELEARIDLISAETIREVCTKYIYNKSPAVAAVGPIGQ 463
Query 193 MP 194
+P
Sbjct 464 LP 465
> dre:503532 pmpcb, zgc:110738; peptidase (mitochondrial processing)
beta (EC:3.4.24.64); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=470
Score = 114 bits (284), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 61/169 (36%), Positives = 91/169 (53%), Gaps = 6/169 (3%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTGL 81
D + M+ +IG++ + G N + + Q F +FNTCY DTGL
Sbjct 293 DTIPLMVANTLIGNWDRSLGG------GMNLSSKLAQMSCQGNLCHSFQSFNTCYTDTGL 346
Query 82 FGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSVAEDL 141
+G Y C+ V + +L SVT+ EV RAK L T +L HLD +T + ED+
Sbjct 347 WGLYMVCEPGTVHDMIRFTQLEWKSLCTSVTESEVNRAKNLLKTNMLLHLDGSTPICEDI 406
Query 142 GRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPL 190
GRQ+L Y R +P E R++AI+A IK V KY+++K A++A+GP+
Sbjct 407 GRQMLCYSRRIPLHELEARIDAINATTIKDVCLKYIYNKAPAIAAVGPI 455
> xla:496289 hypothetical LOC496289; K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=479
Score = 112 bits (281), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 67/184 (36%), Positives = 100/184 (54%), Gaps = 26/184 (14%)
Query 22 DVVAFMLMQAIIGSY-RKHEEGLVPGKLSANTTVRNIATKM-QTGC----AEMFSAFNTC 75
D + M+ +IG++ R G+ N+++K+ Q C F +FNTC
Sbjct 299 DTIPLMVANTLIGNWDRSFGSGV------------NLSSKLAQLTCHGNLCHSFQSFNTC 346
Query 76 YRDTGLFGFYAQCDEVAVEHCVLDLMHGIT----ALSYSVTDEEVERAKAQLTTQLLGHL 131
Y DTGL+G Y C+ VE D+MH + L SVT+ EV RAK L T +L L
Sbjct 347 YTDTGLWGLYMVCEPNTVE----DMMHFVQREWIRLCTSVTENEVARAKNLLKTNMLLQL 402
Query 132 DSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLH 191
D +T + ED+GRQ+L Y R +P E R++ I A+ I+ V KY+++K AV+A+GP+
Sbjct 403 DGSTPICEDIGRQMLCYNRRIPLPELEARIDLISAETIREVCTKYIYNKSPAVAAVGPIG 462
Query 192 GMPQ 195
+P
Sbjct 463 ELPN 466
> xla:379401 uqcrc1, MGC53748; ubiquinol-cytochrome c reductase
core protein I (EC:1.10.2.2); K00414 ubiquinol-cytochrome
c reductase core subunit 1 [EC:1.10.2.2]
Length=478
Score = 108 bits (271), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 66/178 (37%), Positives = 98/178 (55%), Gaps = 14/178 (7%)
Query 21 ADVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTG 80
+D ++ ++ AIIG+Y G GK N + R + + + + FN Y DTG
Sbjct 297 SDNISLLVANAIIGNYDVTYGG---GK---NLSSRVASVAAEHKLCQSYQTFNIRYSDTG 350
Query 81 LFGFYAQCDEVAVEHCVLDLMH----GITALSYSVTDEEVERAKAQLTTQLLGHLDSTTS 136
LFG + D+ H + D++H +L SVTD EV +AK L T L+ LD TT
Sbjct 351 LFGMHFVTDK----HNIEDMLHIAQGEWMSLCTSVTDSEVAQAKNALKTALVAQLDGTTP 406
Query 137 VAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMP 194
V ED+GRQIL YG+ V E R++A+DA ++ + KYL+DK AV+ +GP+ +P
Sbjct 407 VCEDIGRQILSYGQRVSLEELNARIDAVDAKKVSEICSKYLYDKCPAVAGVGPIEQIP 464
> hsa:7384 UQCRC1, D3S3191, QCR1, UQCR1; ubiquinol-cytochrome
c reductase core protein I (EC:1.10.2.2); K00414 ubiquinol-cytochrome
c reductase core subunit 1 [EC:1.10.2.2]
Length=480
Score = 107 bits (268), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 62/173 (35%), Positives = 91/173 (52%), Gaps = 6/173 (3%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTGL 81
D VA + AIIG Y G V LS+ +A K+ + F F+ CY +TGL
Sbjct 300 DNVALQVANAIIGHYDCTYGGGV--HLSSPLASGAVANKL----CQSFQTFSICYAETGL 353
Query 82 FGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSVAEDL 141
G + CD + ++ + L L S T+ EV R K L L+ HLD TT V ED+
Sbjct 354 LGAHFVCDRMKIDDMMFVLQGQWMRLCTSATESEVARGKNILRNALVSHLDGTTPVCEDI 413
Query 142 GRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMP 194
GR +L YGR +P AE+ R+ +DA ++ + KY++D+ AV+ GP+ +P
Sbjct 414 GRSLLTYGRRIPLAEWESRIAEVDASVVREICSKYIYDQCPAVAGYGPIEQLP 466
> hsa:9512 PMPCB, Beta-MPP, MPP11, MPPB, MPPP52, P-52; peptidase
(mitochondrial processing) beta (EC:3.4.24.64); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=489
Score = 107 bits (268), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 60/184 (32%), Positives = 100/184 (54%), Gaps = 24/184 (13%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKM-QTGC----AEMFSAFNTCY 76
D + M+ +IG++ + G + N+++K+ Q C F +FNT Y
Sbjct 309 DTICLMVANTLIGNWDRSFGGGM-----------NLSSKLAQLTCHGNLCHSFQSFNTSY 357
Query 77 RDTGLFGFYAQCDEVAVEHCVLDLMHGITA----LSYSVTDEEVERAKAQLTTQLLGHLD 132
DTGL+G Y C+ V D++H + L SVT+ EV RA+ L T +L LD
Sbjct 358 TDTGLWGLYMVCESSTVA----DMLHVVQKEWMRLCTSVTESEVARARNLLKTNMLLQLD 413
Query 133 STTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHG 192
+T + ED+GRQ+L Y R +P E R++A++A+ I+ V KY++++ A++A+GP+
Sbjct 414 GSTPICEDIGRQMLCYNRRIPIPELEARIDAVNAETIREVCTKYIYNRSPAIAAVGPIKQ 473
Query 193 MPQL 196
+P
Sbjct 474 LPDF 477
> xla:100505438 hypothetical protein LOC100505438; K00414 ubiquinol-cytochrome
c reductase core subunit 1 [EC:1.10.2.2]
Length=480
Score = 106 bits (265), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 63/173 (36%), Positives = 89/173 (51%), Gaps = 6/173 (3%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTGL 81
D V + AIIG Y G V LS+ +A K+ + F FN Y DTGL
Sbjct 300 DNVTLQVANAIIGHYDCTYGGGV--HLSSPLASVAVANKL----CQSFQTFNISYSDTGL 353
Query 82 FGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSVAEDL 141
G + CD ++++ V L L S T+ EV R K L L+ HLD TT V ED+
Sbjct 354 LGAHFVCDAMSIDDMVFFLQGQWMRLCTSATESEVTRGKNILRNALVSHLDGTTPVCEDI 413
Query 142 GRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMP 194
GR +L YGR +P AE+ R+ +DA ++ + KY +D+ AV+ GP+ +P
Sbjct 414 GRSLLTYGRRIPLAEWESRIQEVDAQMLRDICSKYFYDQCPAVAGYGPIEQLP 466
> mmu:22273 Uqcrc1, 1110032G10Rik, MGC97899; ubiquinol-cytochrome
c reductase core protein 1 (EC:1.10.2.2); K00414 ubiquinol-cytochrome
c reductase core subunit 1 [EC:1.10.2.2]
Length=480
Score = 106 bits (265), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 63/173 (36%), Positives = 89/173 (51%), Gaps = 6/173 (3%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTGL 81
D V + AIIG Y G V LS+ +A K+ + F FN Y DTGL
Sbjct 300 DNVTLQVANAIIGHYDCTYGGGV--HLSSPLASVAVANKL----CQSFQTFNISYSDTGL 353
Query 82 FGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSVAEDL 141
G + CD ++++ V L L S T+ EV R K L L+ HLD TT V ED+
Sbjct 354 LGAHFVCDAMSIDDMVFFLQGQWMRLCTSATESEVTRGKNILRNALVSHLDGTTPVCEDI 413
Query 142 GRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMP 194
GR +L YGR +P AE+ R+ +DA ++ + KY +D+ AV+ GP+ +P
Sbjct 414 GRSLLTYGRRIPLAEWESRIQEVDAQMLRDICSKYFYDQCPAVAGYGPIEQLP 466
> dre:393793 uqcrc1, MGC73404, zgc:73404, zgc:85750; ubiquinol-cytochrome
c reductase core protein I (EC:1.10.2.2); K00414
ubiquinol-cytochrome c reductase core subunit 1 [EC:1.10.2.2]
Length=474
Score = 102 bits (253), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 63/182 (34%), Positives = 99/182 (54%), Gaps = 24/182 (13%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEM-----FSAFNTCY 76
D+V M+ +IIGSY G GK ++++++ AE+ F F + Y
Sbjct 294 DIVPLMVANSIIGSYDITFGG---GK--------HLSSRLAQRAAELNLCHSFQTFYSSY 342
Query 77 RDTGLFGFYAQCDEVAVEHCVLDLMH----GITALSYSVTDEEVERAKAQLTTQLLGHLD 132
DTGL G Y +++ +E D+MH + +VT+ +V RAK L L+G L+
Sbjct 343 SDTGLLGIYFVTEKLKIE----DMMHWAQNAWINVCTTVTESDVARAKNALRASLVGQLN 398
Query 133 STTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHG 192
TT V +++GR IL YGR +P AE+ R+ A+ ++ V KY++DK AVSA+GP+
Sbjct 399 GTTPVCDEIGRHILNYGRRIPLAEWDARIEAVTPSVVRDVCSKYIYDKCPAVSAVGPIEQ 458
Query 193 MP 194
+P
Sbjct 459 LP 460
> cel:ZC410.2 mppb-1; Mitochondrial Processing Peptidase Beta
family member (mppb-1); K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=458
Score = 93.2 bits (230), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 51/173 (29%), Positives = 87/173 (50%), Gaps = 6/173 (3%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTGL 81
D +A M+ ++G Y + N R Q E+F +FNTCY++TGL
Sbjct 279 DNLALMVANTLMGEYDRMR------GFGVNAPTRLAEKLSQDAGIEVFQSFNTCYKETGL 332
Query 82 FGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSVAEDL 141
G Y ++++ + ++ L+ ++ + V+RAK L T LL LD +T V ED+
Sbjct 333 VGTYFVAAPESIDNLIDSVLQQWVWLANNIDEAAVDRAKRSLHTNLLLMLDGSTPVCEDI 392
Query 142 GRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMP 194
GRQ+L YGR +PT E R+ +I +++ V + + +V+ + +G P
Sbjct 393 GRQLLCYGRRIPTPELHARIESITVQQLRDVCRRVFLEGQVSAAVVGKTQYWP 445
> sce:YLR163C MAS1, MIF1; Mas1p (EC:3.4.24.64); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=462
Score = 80.5 bits (197), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 59/205 (28%), Positives = 101/205 (49%), Gaps = 19/205 (9%)
Query 3 FIPQFTSLPVSPLLLLL------CADVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRN 56
FI + T LP + + + L D + QAI+G++ + G + + +
Sbjct 257 FIKENT-LPTTHIAIALEGVSWSAPDYFVALATQAIVGNWDR-----AIGTGTNSPSPLA 310
Query 57 IATKMQTGCAEMFSAFNTCYRDTGLFGFYAQCDEVAVEHCVLDLMHGITA-----LSYSV 111
+A A + +F+T Y D+GL+G Y D + EH V +++ I S +
Sbjct 311 VAASQNGSLANSYMSFSTSYADSGLWGMYIVTD--SNEHNVQLIVNEILKEWKRIKSGKI 368
Query 112 TDEEVERAKAQLTTQLLGHLDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKR 171
+D EV RAKAQL LL LD +T++ ED+GRQ++ G+ + E +++ I D+I
Sbjct 369 SDAEVNRAKAQLKAALLLSLDGSTAIVEDIGRQVVTTGKRLSPEEVFEQVDKITKDDIIM 428
Query 172 VAWKYLHDKEVAVSAMGPLHGMPQL 196
A L +K V++ A+G +P +
Sbjct 429 WANYRLQNKPVSMVALGNTSTVPNV 453
> sce:YHR024C MAS2, MIF2; Mas2p (EC:3.4.24.64); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=482
Score = 73.9 bits (180), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 39/107 (36%), Positives = 59/107 (55%), Gaps = 4/107 (3%)
Query 71 AFNTCYRDTGLFGFYAQC----DEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQ 126
AFN Y D+G+FG C AVE + + +T++EV RAK QL +
Sbjct 318 AFNHSYSDSGIFGISLSCIPQAAPQAVEVIAQQMYNTFANKDLRLTEDEVSRAKNQLKSS 377
Query 127 LLGHLDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVA 173
LL +L+S ED+GRQ+L++GR +P E + ++ + D+I RVA
Sbjct 378 LLMNLESKLVELEDMGRQVLMHGRKIPVNEMISKIEDLKPDDISRVA 424
> xla:734517 pmpca, MGC114896; peptidase (mitochondrial processing)
alpha (EC:3.4.24.64); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=518
Score = 72.8 bits (177), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 44/125 (35%), Positives = 69/125 (55%), Gaps = 1/125 (0%)
Query 70 SAFNTCYRDTGLFGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLG 129
++++ Y DTGL +A D V V + T ++ SV + E+ RA+ QL + L+
Sbjct 371 TSYHHSYEDTGLLCIHASADPRQVRDMVEIITREFTLMAGSVGEVELNRARTQLKSMLMM 430
Query 130 HLDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGP 189
+L+S + ED+GRQ+L G E +N + A +IKRVA K L +K AV+A+G
Sbjct 431 NLESRPVIFEDVGRQVLATGTRKLPHELCNLINNVKASDIKRVATKMLRNKP-AVAALGD 489
Query 190 LHGMP 194
L +P
Sbjct 490 LTELP 494
> tgo:TGME49_002680 mitochondrial-processing peptidase alpha subunit,
putative (EC:3.4.24.64); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=563
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 54/175 (30%), Positives = 83/175 (47%), Gaps = 5/175 (2%)
Query 21 ADVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTG 80
D+VA+ ++Q I+G PGK N+ Q E AFNT Y D+G
Sbjct 379 GDLVAYSVLQTILGGGGAFST-GGPGKGMYTRLYLNVLN--QNEWVESAMAFNTQYTDSG 435
Query 81 LFGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSVAED 140
+FG Y D + V + + SVT EE++RAK L + + +L+ V ED
Sbjct 436 IFGLYMLADPTKSANAVKVMAEQFGKM-VSVTKEELQRAKNSLKSSIFMNLECRGIVMED 494
Query 141 LGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMPQ 195
+GRQ+L+ R + EF ++A+ +IKRV ++ K V A G + +P
Sbjct 495 VGRQLLMSNRVISPQEFCTAIDAVTEADIKRVV-DAMYKKPPTVVAYGDVSTVPH 548
> sce:YBL045C COR1, QCR1; Cor1p; K00414 ubiquinol-cytochrome c
reductase core subunit 1 [EC:1.10.2.2]
Length=457
Score = 70.1 bits (170), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 44/134 (32%), Positives = 73/134 (54%), Gaps = 11/134 (8%)
Query 67 EMFSAFNTCYRDTGLFGFYAQCDEVAVEHCVLDLMH----GITALSYSVTDEEVERAKAQ 122
+ F+ F+ Y+D+GL+GF V + + DL+H L+ SVTD EVERAK+
Sbjct 313 DNFNHFSLSYKDSGLWGFSTATRNVTM---IDDLIHFTLKQWNRLTISVTDTEVERAKSL 369
Query 123 LTTQLLGHLDSTTSVAED---LGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHD 179
L Q LG L + + D LG ++L+ G + E +++AI ++K A K L D
Sbjct 370 LKLQ-LGQLYESGNPVNDANLLGAEVLIKGSKLSLGEAFKKIDAITVKDVKAWAGKRLWD 428
Query 180 KEVAVSAMGPLHGM 193
+++A++ G + G+
Sbjct 429 QDIAIAGTGQIEGL 442
> xla:380093 uqcrc2, MGC53943; Ubiquinol-cytochrome C reductase
complex; K00415 ubiquinol-cytochrome c reductase core subunit
2 [EC:1.10.2.2]
Length=451
Score = 69.3 bits (168), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 47/173 (27%), Positives = 85/173 (49%), Gaps = 12/173 (6%)
Query 25 AFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMF--SAFNTCYRDTGLF 82
AF ++Q I+G+ P N T ++ + + F SAFN Y D+GLF
Sbjct 283 AFSVLQHILGAG--------PFIKRGNNTSSKLSQAVNKATNQPFDVSAFNASYSDSGLF 334
Query 83 GFYAQCDEVAVEHCVLDLMHGITALSY-SVTDEEVERAKAQLTTQLLGHLDSTTSVAEDL 141
G Y A + ++ + A++ +VT+ +V RAK QL +Q L L+S+ + D+
Sbjct 335 GVYTVSQAAAASEVINAALNQVKAVAQGNVTEADVTRAKNQLKSQYLMPLESSCGLIGDI 394
Query 142 GRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMP 194
G Q L G + E + +++++ + ++ A K+ K+ +++A G L P
Sbjct 395 GSQALASGTYTTPTETIQQIDSVTSADVVSAAKKFASGKK-SMAATGNLENTP 446
> dre:492801 pmpca, wu:fi19e06, wu:fj83d11, zgc:101647; peptidase
(mitochondrial processing) alpha (EC:3.4.24.64); K01412
mitochondrial processing peptidase [EC:3.4.24.64]
Length=517
Score = 67.4 bits (163), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 40/125 (32%), Positives = 67/125 (53%), Gaps = 1/125 (0%)
Query 70 SAFNTCYRDTGLFGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLG 129
++++ Y D+GL +A D V V + ++ + + E+ERAK QL + L+
Sbjct 370 TSYHHSYEDSGLLCIHASADPRQVREMVEIITREFIQMTGTAGEMELERAKTQLKSMLMM 429
Query 130 HLDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGP 189
+L+S + ED+GRQ+L G+ E ++ + A +IKRV K L K AV+A+G
Sbjct 430 NLESRPVIFEDVGRQVLATGKRKLPHELCELISTVTASDIKRVTMKMLRSKP-AVAALGD 488
Query 190 LHGMP 194
L +P
Sbjct 489 LTELP 493
> xla:446522 uqcrc2, MGC130698, MGC80228; ubiquinol-cytochrome
c reductase core protein II (EC:1.10.2.2); K00415 ubiquinol-cytochrome
c reductase core subunit 2 [EC:1.10.2.2]
Length=451
Score = 67.0 bits (162), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 69/126 (54%), Gaps = 2/126 (1%)
Query 70 SAFNTCYRDTGLFGFYAQCDEVAVEHCVLDLMHGITALSY-SVTDEEVERAKAQLTTQLL 128
SAFN Y D+GLFG Y A + ++ + A++ +VT+ +V RAK QL +Q L
Sbjct 322 SAFNASYSDSGLFGIYTVSQAAAASEVINAALNQVKAVAQGNVTEADVTRAKNQLKSQYL 381
Query 129 GHLDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMG 188
L+S+ + ++G Q L G ++ E + +++++ + ++ A K+ K+ +++A G
Sbjct 382 MTLESSCGLIGEIGSQALASGTYITPTETIQQIDSVTSADVVSAAKKFASGKK-SMAATG 440
Query 189 PLHGMP 194
L P
Sbjct 441 NLENTP 446
> hsa:23203 PMPCA, Alpha-MPP, FLJ26258, INPP5E, KIAA0123, MGC104197;
peptidase (mitochondrial processing) alpha (EC:3.4.24.64);
K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=525
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 43/126 (34%), Positives = 68/126 (53%), Gaps = 3/126 (2%)
Query 70 SAFNTCYRDTGLFGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLG 129
++++ Y DTGL +A D V V + + +V E+ERAK QLT+ L+
Sbjct 378 TSYHHSYEDTGLLCIHASADPRQVREMVEIITKEFILMGGTVDTVELERAKTQLTSMLMM 437
Query 130 HLDSTTSVAEDLGRQIL-VYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMG 188
+L+S + ED+GRQ+L R +P E + + +++KRVA K L K AV+A+G
Sbjct 438 NLESRPVIFEDVGRQVLATRSRKLP-HELCTLIRNVKPEDVKRVASKMLRGKP-AVAALG 495
Query 189 PLHGMP 194
L +P
Sbjct 496 DLTDLP 501
> mmu:66865 Pmpca, 1200002L24Rik, 4933435E07Rik, Alpha-MPP, P-55;
peptidase (mitochondrial processing) alpha (EC:3.4.24.64);
K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=524
Score = 64.3 bits (155), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 43/126 (34%), Positives = 68/126 (53%), Gaps = 3/126 (2%)
Query 70 SAFNTCYRDTGLFGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLG 129
++++ Y DTGL +A D V V + + +V E+ERAK QL + L+
Sbjct 377 TSYHHSYEDTGLLCIHASADPRQVREMVEIITKEFILMGRTVDLVELERAKTQLMSMLMM 436
Query 130 HLDSTTSVAEDLGRQIL-VYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMG 188
+L+S + ED+GRQ+L + R +P E + + ++IKRVA K L K AV+A+G
Sbjct 437 NLESRPVIFEDVGRQVLATHSRKLP-HELCTLIRNVKPEDIKRVASKMLRGKP-AVAALG 494
Query 189 PLHGMP 194
L +P
Sbjct 495 DLTDLP 500
> dre:322549 uqcrc2b, fi06a11, uqcrc2, wu:fb64g04, wu:fi06a11;
ubiquinol-cytochrome c reductase core protein IIb (EC:1.10.2.2);
K00415 ubiquinol-cytochrome c reductase core subunit 2
[EC:1.10.2.2]
Length=454
Score = 59.7 bits (143), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 46/171 (26%), Positives = 84/171 (49%), Gaps = 10/171 (5%)
Query 26 FMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTGLFGFY 85
F ++Q ++G+ + + G S++T+ A T SAFN Y D+GLFG Y
Sbjct 287 FSVLQHVLGAGPRVKRG------SSSTSTLTQAISKVTALPFDASAFNANYTDSGLFGLY 340
Query 86 AQCDEVAVEHCVLDLMHGITALSY-SVTDEEVERAKAQLTTQLLGHLDSTTSVAEDLGRQ 144
C AV + + + A++ ++ ++ +AK QLT L ++S+ + + +G
Sbjct 341 TICQANAVNDVIKAAVGQVNAIAQGNLAAADLSKAKNQLTADYLMSIESSEGLMDVIGTH 400
Query 145 ILVYGR-HVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMP 194
+L G H P A ++NA+ + ++ VA K++ K+ S+ G L P
Sbjct 401 VLSEGTYHTPEA-VTQKINAVSSADVVNVAKKFMSGKKTMASS-GNLVNTP 449
> dre:436930 uqcrc2a, fb33d01, wu:fb33d01, wu:fc43c05, zgc:92453;
ubiquinol-cytochrome c reductase core protein IIa (EC:1.10.2.2);
K00415 ubiquinol-cytochrome c reductase core subunit
2 [EC:1.10.2.2]
Length=460
Score = 59.7 bits (143), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 45/184 (24%), Positives = 90/184 (48%), Gaps = 20/184 (10%)
Query 20 CADVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMF------SAFN 73
A+ AF ++Q I+G+ + G NI++K+ G A+ +AF+
Sbjct 287 SAEANAFSVLQRILGAGPHVKRG------------SNISSKLSQGIAKATAQPFDATAFS 334
Query 74 TCYRDTGLFGFYAQCDEVAVEHCVLDLMHGITALSYS-VTDEEVERAKAQLTTQLLGHLD 132
T Y D+GLFG Y + + + +TA++ +T +++ RAK QL L L+
Sbjct 335 TTYSDSGLFGLYVISQADSTREVISSAVAQVTAVAEGKLTTDDLTRAKNQLKADYLMSLE 394
Query 133 STTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHG 192
S+ + E+LG Q+L G + ++++ + ++ + A +++ ++ ++S+ G L
Sbjct 395 SSDVLLEELGVQLLNSGVYSSPQTVTQSIDSVTSSDVLKAARRFVEGQK-SMSSCGYLEN 453
Query 193 MPQL 196
P L
Sbjct 454 TPFL 457
> bbo:BBOV_III003850 17.m07356; mitochondrial processing peptidase
alpha subunit; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=496
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 50/175 (28%), Positives = 82/175 (46%), Gaps = 4/175 (2%)
Query 21 ADVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTG 80
+++V F ++Q+++G PGK + N+ K + E AF+T Y D G
Sbjct 316 SELVVFTVLQSLLGGGGAFST-GGPGKGMHSRLFLNVLNKHE--FVESCMAFSTVYSDAG 372
Query 81 LFGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSVAED 140
+FG Y A +D+M SVT +E+ERAK L + L L+ ED
Sbjct 373 MFGMYMVVAPQA-SRGAIDVMSNEFRNMLSVTPKELERAKNSLKSFLHMSLEHKAVQMED 431
Query 141 LGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMPQ 195
+ RQ+L+ R + E ++++ A +I+R L + +V A+G L MP
Sbjct 432 IARQLLLCDRVLTVPELERAIDSVTALDIQRCVQSMLKGSKPSVVALGNLAFMPH 486
> mmu:67003 Uqcrc2, 1500004O06Rik; ubiquinol cytochrome c reductase
core protein 2 (EC:1.10.2.2); K00415 ubiquinol-cytochrome
c reductase core subunit 2 [EC:1.10.2.2]
Length=453
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 46/180 (25%), Positives = 90/180 (50%), Gaps = 14/180 (7%)
Query 21 ADVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMF--SAFNTCYRD 78
A+ AF ++Q ++G+ + G N T ++ + G + F SAFN Y D
Sbjct 281 AEANAFSVLQHLLGAGPHIKRG--------NNTTSLLSQSVAKGSHQPFDVSAFNASYSD 332
Query 79 TGLFGFYAQCDEVAVEHCVLDLMHGITALSY-SVTDEEVERAKAQLTTQLLGHLDSTTSV 137
+GLFG Y A + + + A++ +++ +V+ AK +L L ++++
Sbjct 333 SGLFGIYTISQAAAAGEVINAAYNQVKAVAQGNLSSADVQAAKNKLKAGYLMSVETSEGF 392
Query 138 AEDLGRQILVYGRHVPTAEFLMRLNAI-DADEIKRVAWKYLHDKEVAVSAMGPLHGMPQL 196
++G Q L G ++P + L +++++ DAD +K A K++ K+ +++A G L P L
Sbjct 393 LSEIGSQALAAGSYMPPSTVLQQIDSVADADVVK-AAKKFVSGKK-SMAASGNLGHTPFL 450
> ath:AT3G16480 MPPalpha (mitochondrial processing peptidase alpha
subunit); catalytic/ metal ion binding / metalloendopeptidase/
zinc ion binding; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=499
Score = 57.4 bits (137), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 61/126 (48%), Gaps = 2/126 (1%)
Query 70 SAFNTCYRDTGLFGFYA-QCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLL 128
+AF + + +TGLFG Y E A + L V + ++RAKA + +L
Sbjct 365 TAFTSVFNNTGLFGIYGCTSPEFASQGIELVASEMNAVADGKVNQKHLDRAKAATKSAIL 424
Query 129 GHLDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMG 188
+L+S AED+GRQIL YG P +FL ++ + +I K + K + ++ G
Sbjct 425 MNLESRMIAAEDIGRQILTYGERKPVDQFLKTVDQLTLKDIADFTSKVI-TKPLTMATFG 483
Query 189 PLHGMP 194
+ +P
Sbjct 484 DVLNVP 489
> cel:F56D2.1 ucr-1; Ubiquinol-Cytochrome c oxidoReductase complex
family member (ucr-1)
Length=471
Score = 57.4 bits (137), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 34/127 (26%), Positives = 61/127 (48%), Gaps = 3/127 (2%)
Query 72 FNTCYRDTGLFGFYAQCDEVAVEHC---VLDLMHGITALSYSVTDEEVERAKAQLTTQLL 128
FN Y+DTGLFG Y D + + + H L+ + T+EEV AK Q T L
Sbjct 332 FNINYKDTGLFGIYFVADAHDLNDTSGIMKSVAHEWKHLASAATEEEVAMAKNQFRTNLY 391
Query 129 GHLDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMG 188
+L++ T A +++L G +E ++ +DA ++ ++++D+++A +G
Sbjct 392 QNLETNTQKAGFNAKELLYTGNLRQLSELEAQIQKVDAGAVREAISRHVYDRDLAAVGVG 451
Query 189 PLHGMPQ 195
P
Sbjct 452 RTEAFPN 458
> hsa:7385 UQCRC2, QCR2, UQCR2; ubiquinol-cytochrome c reductase
core protein II (EC:1.10.2.2); K00415 ubiquinol-cytochrome
c reductase core subunit 2 [EC:1.10.2.2]
Length=453
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 43/178 (24%), Positives = 89/178 (50%), Gaps = 12/178 (6%)
Query 20 CADVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMF--SAFNTCYR 77
A+ AF ++Q ++G+ + G +NTT ++ + + F SAFN Y
Sbjct 280 SAEANAFSVLQHVLGAGPHVKRG-------SNTT-SHLHQAVAKATQQPFDVSAFNASYS 331
Query 78 DTGLFGFYAQCDEVAVEHCVLDLMHGITALSY-SVTDEEVERAKAQLTTQLLGHLDSTTS 136
D+GLFG Y A + + + ++ ++++ +V+ AK +L L ++S+
Sbjct 332 DSGLFGIYTISQATAAGDVIKAAYNQVKTIAQGNLSNTDVQAAKNKLKAGYLMSVESSEC 391
Query 137 VAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMP 194
E++G Q LV G ++P + L +++++ +I A K++ ++ +++A G L P
Sbjct 392 FLEEVGSQALVAGSYMPPSTVLQQIDSVANADIINAAKKFVSGQK-SMAASGNLGHTP 448
> pfa:PFE1155c mitochondrial processing peptidase alpha subunit,
putative; K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=534
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 35/125 (28%), Positives = 62/125 (49%), Gaps = 2/125 (1%)
Query 71 AFNTCYRDTGLFGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGH 130
AF+T + DTGLFG Y + + + ++ VTDEE+ RAK L + +
Sbjct 401 AFSTQHSDTGLFGLYFTGEPSNTSDIIKAMALEFQKMN-RVTDEELNRAKKSLKSFMWMS 459
Query 131 LDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPL 190
L+ + + EDL RQ+++ R + + +++I ++I+RV +L K V G +
Sbjct 460 LEYKSILMEDLARQMMILNRILTGKQLSDAIDSITKEDIQRVVHNFLKTKPTVV-VYGNI 518
Query 191 HGMPQ 195
+ P
Sbjct 519 NYSPH 523
> cel:Y71G12B.24 mppa-1; Mitochondrial Processing Peptidase Alpha
family member (mppa-1); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=477
Score = 50.8 bits (120), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/110 (28%), Positives = 53/110 (48%), Gaps = 0/110 (0%)
Query 71 AFNTCYRDTGLFGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGH 130
A N Y D+G+F A + ++ L+H I L V E+ RA+ QL + L+ +
Sbjct 336 AHNHSYSDSGVFTVTASSPPENINDALILLVHQILQLQQGVEPTELARARTQLRSHLMMN 395
Query 131 LDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDK 180
L+ + ED+ RQ+L +G E+ ++ + +I RV + L K
Sbjct 396 LEVRPVLFEDMVRQVLGHGDRKQPEEYAEKIEKVTNSDIIRVTERLLASK 445
> ath:AT1G51980 mitochondrial processing peptidase alpha subunit,
putative; K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=451
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 31/84 (36%), Positives = 43/84 (51%), Gaps = 7/84 (8%)
Query 70 SAFNTCYRDTGLFGFYA----QCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTT 125
+AF + + DTGLFG Y Q A+E +L V ++RAKA +
Sbjct 369 TAFTSIFNDTGLFGIYGCSSPQFAAKAIELAAKELKD---VAGGKVNQAHLDRAKAATKS 425
Query 126 QLLGHLDSTTSVAEDLGRQILVYG 149
+L +L+S AED+GRQIL YG
Sbjct 426 AVLMNLESRMIAAEDIGRQILTYG 449
> tpv:TP02_0218 ubiquinol-cytochrome C reductase complex core
protein II, mitochondrial precursor (EC:1.10.2.2); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=525
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 33/128 (25%), Positives = 56/128 (43%), Gaps = 3/128 (2%)
Query 71 AFNTCYRDTGLFGFYAQCDEVAVE---HCVLDLMHGITALSYSVTDEEVERAKAQLTTQL 127
AFNT + +GLFG Y + V L+ +T+ E+ K L + L
Sbjct 389 AFNTVHSTSGLFGIYLVVNGAYASGNMDQVFTLVRDEFERMKKITNHELSGGKNSLKSFL 448
Query 128 LGHLDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAM 187
L+ V ED+GRQ+L R + ++ ++ + D+IK V + ++ +V
Sbjct 449 HMSLEHKAVVCEDVGRQLLFCNRVLDPSDLENLIDEVTLDDIKAVVNELRVNQTPSVVVY 508
Query 188 GPLHGMPQ 195
G L +P
Sbjct 509 GKLSRVPH 516
> cpv:cgd7_2080 mitochondrial processing peptidase, insulinase
like metalloprotease ; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=497
Score = 38.5 bits (88), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 28/110 (25%), Positives = 51/110 (46%), Gaps = 13/110 (11%)
Query 70 SAFNTCYRDTGLFGFYA------QCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQL 123
+ F Y DTGLFG + + + V L M I+ + E+ERAK +
Sbjct 356 NCFVNQYSDTGLFGIHITSYPGYSLESIKVIAKQLGKMKNIS-------ERELERAKNLV 408
Query 124 TTQLLGHLDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVA 173
+ + ++ + E++ +QIL Y + E + + +I ++IK+VA
Sbjct 409 LSTICTAYENRSHYMEEISKQILSYSEFIELDEIINCIKSIGIEDIKKVA 458
> cpv:cgd5_3400 mitochondrial processing peptidase beta subunit
; K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=375
Score = 36.6 bits (83), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 30/61 (49%), Gaps = 3/61 (4%)
Query 20 CADVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDT 79
D + M +Q+++G Y + V G N + I + ++ E F FNTCY+DT
Sbjct 310 SKDFLKVMFLQSMLGEYGTNNINRVTG--YKNQIIERILSGIKD-HVEFFETFNTCYKDT 366
Query 80 G 80
G
Sbjct 367 G 367
> mmu:74194 Rnd3, 2610017M01Rik, AI661404, Arhe, Rhoe; Rho family
GTPase 3; K07859 Rho family GTPase 3
Length=244
Score = 35.8 bits (81), Expect = 0.100, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 34/62 (54%), Gaps = 3/62 (4%)
Query 85 YAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDST---TSVAEDL 141
Y +C + E+ V D+ H T + T++ V+R K+Q T+ + H+ S ++VA DL
Sbjct 174 YIECSALQSENSVRDIFHVATLACVNKTNKNVKRNKSQRATKRISHMPSRPELSAVATDL 233
Query 142 GR 143
+
Sbjct 234 RK 235
> hsa:390 RND3, ARHE, Rho8, RhoE, memB; Rho family GTPase 3; K07859
Rho family GTPase 3
Length=244
Score = 35.8 bits (81), Expect = 0.100, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 34/62 (54%), Gaps = 3/62 (4%)
Query 85 YAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDST---TSVAEDL 141
Y +C + E+ V D+ H T + T++ V+R K+Q T+ + H+ S ++VA DL
Sbjct 174 YIECSALQSENSVRDIFHVATLACVNKTNKNVKRNKSQRATKRISHMPSRPELSAVATDL 233
Query 142 GR 143
+
Sbjct 234 RK 235
> dre:568464 zfpm2b, FOG-B, fog2b; zinc finger protein, multitype
2b
Length=1093
Score = 32.3 bits (72), Expect = 1.1, Method: Composition-based stats.
Identities = 16/59 (27%), Positives = 29/59 (49%), Gaps = 0/59 (0%)
Query 4 IPQFTSLPVSPLLLLLCADVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQ 62
+PQF + PVSP L+ + V + + Y KH++ PG + + + N + K +
Sbjct 807 VPQFGNTPVSPKGLMDYHECVVCKISFNKVEDYLKHKQNFCPGTILQSASTENKSIKKE 865
> mmu:67453 Slc25a46, 1200007B05Rik, AI325987; solute carrier
family 25, member 46
Length=418
Score = 31.6 bits (70), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 24/92 (26%), Positives = 42/92 (45%), Gaps = 5/92 (5%)
Query 19 LCADVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRD 78
LC+DV+ + L + +R H +G + NT + + T M NT ++
Sbjct 325 LCSDVILYPLETVL---HRLHIQGTR--TIIDNTDLGYEVLPINTQYEGMRDCINTIKQE 379
Query 79 TGLFGFYAQCDEVAVEHCVLDLMHGITALSYS 110
G+FGFY V +++ + + IT + YS
Sbjct 380 EGVFGFYKGFGAVIIQYTLHATILQITKMIYS 411
> dre:569256 MGC153498; zgc:153498
Length=847
Score = 31.6 bits (70), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 24/47 (51%), Gaps = 0/47 (0%)
Query 147 VYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGM 193
V R +P E + ++AID DE++ V +K L E + P G+
Sbjct 592 VVSRELPVGEVIATISAIDIDELELVKYKILSGNERGYFELNPDSGV 638
> hsa:91137 SLC25A46; solute carrier family 25, member 46
Length=418
Score = 31.6 bits (70), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 24/92 (26%), Positives = 42/92 (45%), Gaps = 5/92 (5%)
Query 19 LCADVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRD 78
LC+DV+ + L + +R H +G + NT + + T M NT ++
Sbjct 325 LCSDVILYPLETVL---HRLHIQGTR--TIIDNTDLGYEVLPINTQYEGMRDCINTIRQE 379
Query 79 TGLFGFYAQCDEVAVEHCVLDLMHGITALSYS 110
G+FGFY V +++ + + IT + YS
Sbjct 380 EGVFGFYKGFGAVIIQYTLHAAVLQITKIIYS 411
> xla:432247 slc25a46-a, MGC81360; solute carrier family 25, member
46; K03454 solute carrier family 25, member 46
Length=417
Score = 31.2 bits (69), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 42/92 (45%), Gaps = 5/92 (5%)
Query 19 LCADVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRD 78
LCADV+ + L + +R H +G + NT + + + T + NT R+
Sbjct 324 LCADVLLYPLETVL---HRLHIQGTR--TIIDNTDLGHEVVPINTQYEGLKDCINTIKRE 378
Query 79 TGLFGFYAQCDEVAVEHCVLDLMHGITALSYS 110
G GFY V V++ + ++ IT + YS
Sbjct 379 EGGLGFYKGFGAVVVQYTLHAIVLQITKIIYS 410
> xla:494746 slc25a46-b, slc25a46; solute carrier family 25, member
46; K03454 solute carrier family 25, member 46
Length=414
Score = 31.2 bits (69), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 42/92 (45%), Gaps = 5/92 (5%)
Query 19 LCADVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRD 78
LCADV+ + L + +R H +G + NT + + + T + NT R+
Sbjct 321 LCADVLLYPLETVM---HRLHIQGTR--TIIDNTDLGHEVVPINTQYEGLKDCINTIKRE 375
Query 79 TGLFGFYAQCDEVAVEHCVLDLMHGITALSYS 110
G GFY V V++ + ++ IT + YS
Sbjct 376 EGGLGFYKGFGAVVVQYTLHAIVLQITKIIYS 407
> dre:436831 slc25a46, zgc:92767; solute carrier family 25, member
46
Length=405
Score = 29.3 bits (64), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 39/92 (42%), Gaps = 5/92 (5%)
Query 19 LCADVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRD 78
LCADV+ F L + +R H +G + NT + + T M N R+
Sbjct 313 LCADVLLFPLETVL---HRLHIQGTR--TIIDNTDLGFEVLPINTQYEGMRDCINAIRRE 367
Query 79 TGLFGFYAQCDEVAVEHCVLDLMHGITALSYS 110
G GFY + V++ + + IT + YS
Sbjct 368 EGTMGFYKGFGSIVVQYSLHATVLQITKMIYS 399
Lambda K H
0.323 0.136 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5802328440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40