bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3684_orf1
Length=210
Score E
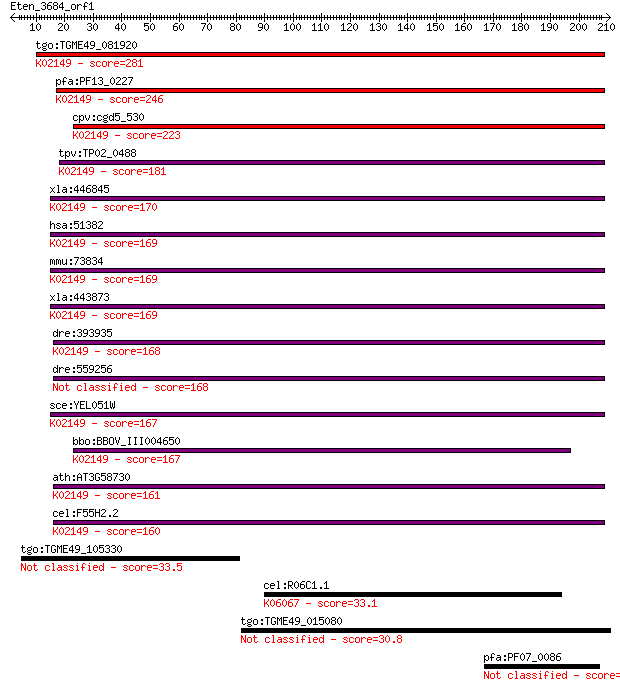
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_081920 vacuolar ATP synthase subunit D, putative (E... 281 8e-76
pfa:PF13_0227 vacuolar ATP synthase subunit D, putative (EC:3.... 246 6e-65
cpv:cgd5_530 vacuolar H-ATpase subunit D ; K02149 V-type H+-tr... 223 4e-58
tpv:TP02_0488 vacuolar ATP synthase subunit D; K02149 V-type H... 181 1e-45
xla:446845 atp6v1d, MGC80692; ATPase, H+ transporting, lysosom... 170 4e-42
hsa:51382 ATP6V1D, ATP6M, VATD, VMA8; ATPase, H+ transporting,... 169 5e-42
mmu:73834 Atp6v1d, 1110004P10Rik, Atp6m, VATD, Vma8; ATPase, H... 169 5e-42
xla:443873 MGC79146 protein; K02149 V-type H+-transporting ATP... 169 8e-42
dre:393935 atp6v1d, MGC55524, zgc:55524; ATPase, H+ transporti... 168 2e-41
dre:559256 atp6v1d; ATPase, H+ transporting, V1 subunit D 168 2e-41
sce:YEL051W VMA8; Subunit D of the eight-subunit V1 peripheral... 167 2e-41
bbo:BBOV_III004650 17.m07416; V-type ATPase, D subunit family ... 167 3e-41
ath:AT3G58730 vacuolar ATP synthase subunit D (VATD) / V-ATPas... 161 1e-39
cel:F55H2.2 vha-14; Vacuolar H ATPase family member (vha-14); ... 160 3e-39
tgo:TGME49_105330 cyclin, N-terminal domain-containing protein... 33.5 0.55
cel:R06C1.1 hda-3; Histone DeAcetylase family member (hda-3); ... 33.1 0.65
tgo:TGME49_015080 hypothetical protein 30.8 3.1
pfa:PF07_0086 conserved Plasmodium membrane protein, unknown f... 30.8 3.3
> tgo:TGME49_081920 vacuolar ATP synthase subunit D, putative
(EC:3.6.3.14); K02149 V-type H+-transporting ATPase subunit
D [EC:3.6.3.14]
Length=245
Score = 281 bits (720), Expect = 8e-76, Method: Compositional matrix adjust.
Identities = 131/199 (65%), Positives = 164/199 (82%), Gaps = 1/199 (0%)
Query 10 MSSDAQVAPSRMTLQVAKQKKKGASQGYQLLKKKSDALSARFRGMLKEIVKTKLSIGDTI 69
M D + AP+RM L + KQK+KGA QGYQLLKKKSDAL+ARFRGMLK+IV+ K ++G +
Sbjct 1 MPQDIRPAPNRMNLAIMKQKRKGAQQGYQLLKKKSDALTARFRGMLKQIVQLKQAVGADM 60
Query 70 NEAHFSMAKASWAGGNDLREQLMQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILK 129
N A FS+AKA+WA G D + QL++R++RPA F+ A DNVAGV LP+F I TDP+VD+LK
Sbjct 61 NTAAFSLAKATWAAG-DFKSQLLERVRRPATFLNVAADNVAGVTLPIFHICTDPSVDVLK 119
Query 130 NINLSAGGHVILAARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKL 189
N+ ++AGG VI+AAR+ + + +ELVKLASLQTAFFTLD EIKMTNRRVNAL+NVVLP++
Sbjct 120 NVGVAAGGQVIMAAREMFLKVFSELVKLASLQTAFFTLDEEIKMTNRRVNALSNVVLPRI 179
Query 190 DKSINYITKELDEMEREEF 208
D INYI +ELDEMEREEF
Sbjct 180 DGGINYIVRELDEMEREEF 198
> pfa:PF13_0227 vacuolar ATP synthase subunit D, putative (EC:3.6.3.14);
K02149 V-type H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=247
Score = 246 bits (627), Expect = 6e-65, Method: Compositional matrix adjust.
Identities = 118/192 (61%), Positives = 149/192 (77%), Gaps = 1/192 (0%)
Query 17 APSRMTLQVAKQKKKGASQGYQLLKKKSDALSARFRGMLKEIVKTKLSIGDTINEAHFSM 76
PSR+TLQ+ KQKKK A QGY LLKKKSDAL FR +LK+IVKTK +G+ + A FS+
Sbjct 10 VPSRITLQLMKQKKKSAFQGYSLLKKKSDALFIHFRDVLKDIVKTKTKVGEEMRNASFSL 69
Query 77 AKASWAGGNDLREQLMQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAG 136
AK+ WA G D + Q+++ IKRP V ++ + +NVAGV+LP+FQ+ DPTVD+L N+ ++AG
Sbjct 70 AKSVWAAG-DFKGQIIEGIKRPVVTLSLSTNNVAGVKLPIFQVNIDPTVDVLGNLGVAAG 128
Query 137 GHVILAARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYI 196
G VI R+ Y + L LVKLAS+Q AFF+LD EIKMTNRRVNALNN+VLP+LD INYI
Sbjct 129 GQVINNTRENYLQCLNMLVKLASMQVAFFSLDEEIKMTNRRVNALNNIVLPRLDGGINYI 188
Query 197 TKELDEMEREEF 208
KELDE+EREEF
Sbjct 189 IKELDEIEREEF 200
> cpv:cgd5_530 vacuolar H-ATpase subunit D ; K02149 V-type H+-transporting
ATPase subunit D [EC:3.6.3.14]
Length=249
Score = 223 bits (568), Expect = 4e-58, Method: Compositional matrix adjust.
Identities = 110/187 (58%), Positives = 142/187 (75%), Gaps = 2/187 (1%)
Query 23 LQVAKQKKKGASQGYQLLKKKSDALSARFRGMLKEIVKTKLSIGDTINEAHFSMAKASWA 82
LQ K K KGA QGY LLK+KSDALS +FRGMLKEIV+TK SIG+ I EA F++AKA+WA
Sbjct 7 LQAIKLKSKGAKQGYDLLKRKSDALSNKFRGMLKEIVETKRSIGNDIKEASFALAKATWA 66
Query 83 GGNDLREQLMQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDI-LKNINLSAGGHVIL 141
G D ++++++ KRP V + +N+AGVRLP+F++ D +I +++GG VI
Sbjct 67 AG-DFKDRIIESCKRPTVTMEVGTENIAGVRLPIFEMNVDNNSSTETCHIGVASGGQVIQ 125
Query 142 AARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELD 201
+ R+ Y + L +LVKLASLQTAFF+LD EIKMTNRRVNAL NVVLPKL+ +NYI +ELD
Sbjct 126 STREIYMKVLRDLVKLASLQTAFFSLDEEIKMTNRRVNALQNVVLPKLEDGMNYILRELD 185
Query 202 EMEREEF 208
E+EREEF
Sbjct 186 EIEREEF 192
> tpv:TP02_0488 vacuolar ATP synthase subunit D; K02149 V-type
H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=238
Score = 181 bits (460), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 89/193 (46%), Positives = 135/193 (69%), Gaps = 3/193 (1%)
Query 18 PSRM--TLQVAKQKKKGASQGYQLLKKKSDALSARFRGMLKEIVKTKLSIGDTINEAHFS 75
PSRM LQ KQ++ A GY LLK+KSDAL+++F +L+ V+ K + + + +A +S
Sbjct 10 PSRMLVNLQNLKQRRHNAHLGYSLLKRKSDALTSKFHRLLRATVQGKERLVEGLKDATYS 69
Query 76 MAKASWAGGNDLREQLMQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSA 135
+A A W+ D + +++ + RP+V + +N+AGV LPVF + TDPTVD+ N++LS+
Sbjct 70 LANAVWSA-EDFKSLVIESVGRPSVTLKLRGENIAGVLLPVFSLQTDPTVDLFANLSLSS 128
Query 136 GGHVILAARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINY 195
GG I + + + AL LV+LASLQ +F L+ EI+MTNRR+NAL+NV++P +D+++ Y
Sbjct 129 GGSAIQSVKTTHLAALDILVELASLQISFIILNEEIRMTNRRINALDNVLIPSIDRNLEY 188
Query 196 ITKELDEMEREEF 208
I +ELDEMEREEF
Sbjct 189 IRRELDEMEREEF 201
> xla:446845 atp6v1d, MGC80692; ATPase, H+ transporting, lysosomal
34kDa, V1 subunit D (EC:3.6.3.14); K02149 V-type H+-transporting
ATPase subunit D [EC:3.6.3.14]
Length=248
Score = 170 bits (430), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 91/194 (46%), Positives = 131/194 (67%), Gaps = 2/194 (1%)
Query 15 QVAPSRMTLQVAKQKKKGASQGYQLLKKKSDALSARFRGMLKEIVKTKLSIGDTINEAHF 74
+V PSRM + K + KGA G LLKKKSDAL+ RFR +LK+I++TK+ +G+ + EA F
Sbjct 8 EVFPSRMAQTIMKARLKGAQTGRNLLKKKSDALTMRFRQILKKIIETKMLMGEVMREAAF 67
Query 75 SMAKASWAGGNDLREQLMQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLS 134
S+A+A + G D ++Q + + V V A DNVAGV LPVF+ + D + L+
Sbjct 68 SLAEAKFTAG-DFSTTVIQNVNKSQVKVRAKKDNVAGVTLPVFEHYQEGG-DSYELTGLA 125
Query 135 AGGHVILAARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSIN 194
GG + + Y +A+ LV+LASLQT+F TLD IK+TNRRVNA+ +V++PK++++++
Sbjct 126 RGGEQLAKLKRNYAKAVELLVELASLQTSFVTLDEAIKITNRRVNAIEHVIIPKIERTLS 185
Query 195 YITKELDEMEREEF 208
YI ELDE EREEF
Sbjct 186 YIITELDEREREEF 199
> hsa:51382 ATP6V1D, ATP6M, VATD, VMA8; ATPase, H+ transporting,
lysosomal 34kDa, V1 subunit D (EC:3.6.3.14 3.6.1.34); K02149
V-type H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=247
Score = 169 bits (429), Expect = 5e-42, Method: Compositional matrix adjust.
Identities = 89/194 (45%), Positives = 131/194 (67%), Gaps = 2/194 (1%)
Query 15 QVAPSRMTLQVAKQKKKGASQGYQLLKKKSDALSARFRGMLKEIVKTKLSIGDTINEAHF 74
++ PSRM + K + KGA G LLKKKSDAL+ RFR +LK+I++TK+ +G+ + EA F
Sbjct 8 EIFPSRMAQTIMKARLKGAQTGRNLLKKKSDALTLRFRQILKKIIETKMLMGEVMREAAF 67
Query 75 SMAKASWAGGNDLREQLMQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLS 134
S+A+A + G D ++Q + + V + A DNVAGV LPVF+ + T D + L+
Sbjct 68 SLAEAKFTAG-DFSTTVIQNVNKAQVKIRAKKDNVAGVTLPVFEHYHEGT-DSYELTGLA 125
Query 135 AGGHVILAARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSIN 194
GG + + Y +A+ LV+LASLQT+F TLD IK+TNRRVNA+ +V++P++++++
Sbjct 126 RGGEQLAKLKRNYAKAVELLVELASLQTSFVTLDEAIKITNRRVNAIEHVIIPRIERTLA 185
Query 195 YITKELDEMEREEF 208
YI ELDE EREEF
Sbjct 186 YIITELDEREREEF 199
> mmu:73834 Atp6v1d, 1110004P10Rik, Atp6m, VATD, Vma8; ATPase,
H+ transporting, lysosomal V1 subunit D; K02149 V-type H+-transporting
ATPase subunit D [EC:3.6.3.14]
Length=247
Score = 169 bits (429), Expect = 5e-42, Method: Compositional matrix adjust.
Identities = 89/194 (45%), Positives = 131/194 (67%), Gaps = 2/194 (1%)
Query 15 QVAPSRMTLQVAKQKKKGASQGYQLLKKKSDALSARFRGMLKEIVKTKLSIGDTINEAHF 74
++ PSRM + K + KGA G LLKKKSDAL+ RFR +LK+I++TK+ +G+ + EA F
Sbjct 8 EIFPSRMAQTIMKARLKGAQTGRNLLKKKSDALTLRFRQILKKIIETKMLMGEVMREAAF 67
Query 75 SMAKASWAGGNDLREQLMQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLS 134
S+A+A + G D ++Q + + V + A DNVAGV LPVF+ + T D + L+
Sbjct 68 SLAEAKFTAG-DFSTTVIQNVNKAQVKIRAKKDNVAGVTLPVFEHYHEGT-DSYELTGLA 125
Query 135 AGGHVILAARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSIN 194
GG + + Y +A+ LV+LASLQT+F TLD IK+TNRRVNA+ +V++P++++++
Sbjct 126 RGGEQLAKLKRNYAKAVELLVELASLQTSFVTLDEAIKITNRRVNAIEHVIIPRIERTLA 185
Query 195 YITKELDEMEREEF 208
YI ELDE EREEF
Sbjct 186 YIITELDEREREEF 199
> xla:443873 MGC79146 protein; K02149 V-type H+-transporting ATPase
subunit D [EC:3.6.3.14]
Length=246
Score = 169 bits (427), Expect = 8e-42, Method: Compositional matrix adjust.
Identities = 90/194 (46%), Positives = 131/194 (67%), Gaps = 2/194 (1%)
Query 15 QVAPSRMTLQVAKQKKKGASQGYQLLKKKSDALSARFRGMLKEIVKTKLSIGDTINEAHF 74
+V PSRM + K + KGA G LLKKKSDAL+ RFR +LK+I++TK+ +G+ + EA F
Sbjct 8 EVFPSRMAQSIMKARLKGAQTGRNLLKKKSDALTMRFRQILKKIIETKMLMGEVMREAAF 67
Query 75 SMAKASWAGGNDLREQLMQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLS 134
S+A+A + G D ++Q + + V V A DNVAGV LPVF+ + D + L+
Sbjct 68 SLAEAKFTAG-DFSTTVIQNVNKAQVKVRARKDNVAGVTLPVFEHYQEGG-DSYELTGLA 125
Query 135 AGGHVILAARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSIN 194
GG + + Y +A+ LV+LASLQT+F TLD IK+TNRRVNA+ +V++P+++++++
Sbjct 126 RGGEQLAKLKRNYAKAVELLVELASLQTSFVTLDEAIKITNRRVNAIEHVIIPRIERTLS 185
Query 195 YITKELDEMEREEF 208
YI ELDE EREEF
Sbjct 186 YIVTELDEREREEF 199
> dre:393935 atp6v1d, MGC55524, zgc:55524; ATPase, H+ transporting,
V1 subunit D; K02149 V-type H+-transporting ATPase subunit
D [EC:3.6.3.14]
Length=248
Score = 168 bits (425), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 90/193 (46%), Positives = 129/193 (66%), Gaps = 2/193 (1%)
Query 16 VAPSRMTLQVAKQKKKGASQGYQLLKKKSDALSARFRGMLKEIVKTKLSIGDTINEAHFS 75
+ PSRM + K + KGA G LLKKKSDALS RFR +L++I++TK +G+ + EA FS
Sbjct 9 IFPSRMAQTIMKARLKGAQTGRSLLKKKSDALSMRFRQILRKIIETKTLMGEVMREAAFS 68
Query 76 MAKASWAGGNDLREQLMQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSA 135
+A+A +A G D ++Q + + V V A DNVAGV LPVF+ + D + L+
Sbjct 69 LAEAKFAAG-DFSTTVIQNVNKAQVKVRAKKDNVAGVTLPVFEHYQEGG-DSYELTGLAR 126
Query 136 GGHVILAARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINY 195
GG + + Y +A+ LV+LASLQT+F TLD IK+TNRRVNA+ +V++P++++++ Y
Sbjct 127 GGEQLSRLKRNYAKAVELLVELASLQTSFVTLDEAIKITNRRVNAIEHVIIPRIERTLTY 186
Query 196 ITKELDEMEREEF 208
I ELDE EREEF
Sbjct 187 IITELDEREREEF 199
> dre:559256 atp6v1d; ATPase, H+ transporting, V1 subunit D
Length=248
Score = 168 bits (425), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 90/193 (46%), Positives = 129/193 (66%), Gaps = 2/193 (1%)
Query 16 VAPSRMTLQVAKQKKKGASQGYQLLKKKSDALSARFRGMLKEIVKTKLSIGDTINEAHFS 75
+ PSRM + K + KGA G LLKKKSDALS RFR +L++I++TK +G+ + EA FS
Sbjct 9 IFPSRMAQTIMKARLKGAQTGRSLLKKKSDALSMRFRQILRKIIETKTLMGEVMREAAFS 68
Query 76 MAKASWAGGNDLREQLMQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSA 135
+A+A +A G D ++Q + + V V A DNVAGV LPVF+ + D + L+
Sbjct 69 LAEAKFAAG-DFSTTVIQNVNKAQVKVRAKKDNVAGVTLPVFEHYQEGG-DSYELTGLAR 126
Query 136 GGHVILAARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINY 195
GG + + Y +A+ LV+LASLQT+F TLD IK+TNRRVNA+ +V++P++++++ Y
Sbjct 127 GGEQLSRLKRNYAKAVELLVELASLQTSFVTLDEAIKITNRRVNAIEHVIIPRIERTLTY 186
Query 196 ITKELDEMEREEF 208
I ELDE EREEF
Sbjct 187 IITELDEREREEF 199
> sce:YEL051W VMA8; Subunit D of the eight-subunit V1 peripheral
membrane domain of the vacuolar H+-ATPase (V-ATPase), an
electrogenic proton pump found throughout the endomembrane system;
plays a role in the coupling of proton transport and
ATP hydrolysis (EC:3.6.3.14); K02149 V-type H+-transporting
ATPase subunit D [EC:3.6.3.14]
Length=256
Score = 167 bits (424), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 87/194 (44%), Positives = 125/194 (64%), Gaps = 0/194 (0%)
Query 15 QVAPSRMTLQVAKQKKKGASQGYQLLKKKSDALSARFRGMLKEIVKTKLSIGDTINEAHF 74
QV P+RMTL + K K KGA+QGY LLK+KS+AL+ RFR + K I K +G + A F
Sbjct 7 QVFPTRMTLGLMKTKLKGANQGYSLLKRKSEALTKRFRDITKRIDDAKQKMGRVMQTAAF 66
Query 75 SMAKASWAGGNDLREQLMQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLS 134
S+A+ S+A G ++ Q+ + + V A +NV+GV L F+ DP ++ + L
Sbjct 67 SLAEVSYATGENIGYQVQESVSTARFKVRARQENVSGVYLSQFESYIDPEINDFRLTGLG 126
Query 135 AGGHVILAARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSIN 194
GG + A++ Y A+ LV+LASLQTAF LD IK+TNRRVNA+ +V++P+ + +I
Sbjct 127 RGGQQVQRAKEIYSRAVETLVELASLQTAFIILDEVIKVTNRRVNAIEHVIIPRTENTIA 186
Query 195 YITKELDEMEREEF 208
YI ELDE++REEF
Sbjct 187 YINSELDELDREEF 200
> bbo:BBOV_III004650 17.m07416; V-type ATPase, D subunit family
protein; K02149 V-type H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=233
Score = 167 bits (423), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 79/174 (45%), Positives = 121/174 (69%), Gaps = 1/174 (0%)
Query 23 LQVAKQKKKGASQGYQLLKKKSDALSARFRGMLKEIVKTKLSIGDTINEAHFSMAKASWA 82
LQ+ KQK+ A GY LLK+KSDAL+++FR +LK+ ++ K + + NEA ++++ A W+
Sbjct 2 LQILKQKRTNAHLGYSLLKRKSDALASKFRKLLKDTIQGKEKVIEGFNEASYALSNAVWS 61
Query 83 GGNDLREQLMQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILA 142
G D + +++ + R AV + +NVAGV +P F++ DPTVD++ NI L+ GGHVI +
Sbjct 62 AG-DFKSLVVESVGRSAVTLRVRTENVAGVIIPHFELKIDPTVDVIANIGLTTGGHVIHS 120
Query 143 ARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYI 196
+ + E L L +LASLQ +F L+ EIKMTNRRVNAL+N+V+P +D ++ YI
Sbjct 121 VKTAHLEFLETLAELASLQVSFMMLEQEIKMTNRRVNALDNLVIPTIDNNLEYI 174
> ath:AT3G58730 vacuolar ATP synthase subunit D (VATD) / V-ATPase
D subunit / vacuolar proton pump D subunit (VATPD); K02149
V-type H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=261
Score = 161 bits (408), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 86/194 (44%), Positives = 128/194 (65%), Gaps = 3/194 (1%)
Query 16 VAPSRMTLQVAKQKKKGASQGYQLLKKKSDALSARFRGMLKEIVKTKLSIGDTINEAHFS 75
V P+ L V K + GA++G+ LLKKKSDAL+ +FR +LK+IV K S+GD + + F+
Sbjct 10 VVPTVTMLGVMKARLVGATRGHALLKKKSDALTVQFRALLKKIVTAKESMGDMMKTSSFA 69
Query 76 MAKASWAGGNDLREQLMQRIKRPAVFVTAAYDNVAGVRLPVF-QITTDPTVDILKNINLS 134
+ + + G++++ +++ +K + V + +N+AGV+LP F + T + L L+
Sbjct 70 LTEVKYVAGDNVKHVVLENVKEATLKVRSRTENIAGVKLPKFDHFSEGETKNDL--TGLA 127
Query 135 AGGHVILAARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSIN 194
GG + A R Y +A+ LV+LASLQT+F TLD IK TNRRVNAL NVV PKL+ +I+
Sbjct 128 RGGQQVRACRVAYVKAIEVLVELASLQTSFLTLDEAIKTTNRRVNALENVVKPKLENTIS 187
Query 195 YITKELDEMEREEF 208
YI ELDE+ERE+F
Sbjct 188 YIKGELDELEREDF 201
> cel:F55H2.2 vha-14; Vacuolar H ATPase family member (vha-14);
K02149 V-type H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=257
Score = 160 bits (405), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 86/193 (44%), Positives = 123/193 (63%), Gaps = 2/193 (1%)
Query 16 VAPSRMTLQVAKQKKKGASQGYQLLKKKSDALSARFRGMLKEIVKTKLSIGDTINEAHFS 75
V PSRM + K + KGA +G+ LLKKK+DAL+ RFR +L++IV+ K+ +G+ + EA FS
Sbjct 11 VFPSRMAQTLMKTRLKGAQKGHSLLKKKADALNLRFRDILRKIVENKVLMGEVMKEAAFS 70
Query 76 MAKASWAGGNDLREQLMQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSA 135
+A+A + G D ++Q + + V +NV GV LPVF D D L
Sbjct 71 LAEAKFTAG-DFSHTVIQNVSQAQYRVRMKKENVVGVFLPVFDAYQDGP-DAYDLTGLGK 128
Query 136 GGHVILAARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINY 195
GG I + Y +A+ LV+LA+LQT F TLD IK+TNRRVNA+ +V++P+++ ++ Y
Sbjct 129 GGANIARLKKNYNKAIELLVELATLQTCFITLDEAIKVTNRRVNAIEHVIIPRIENTLTY 188
Query 196 ITKELDEMEREEF 208
I ELDEMEREEF
Sbjct 189 IVTELDEMEREEF 201
> tgo:TGME49_105330 cyclin, N-terminal domain-containing protein
(EC:5.4.99.17 1.3.1.28)
Length=1174
Score = 33.5 bits (75), Expect = 0.55, Method: Composition-based stats.
Identities = 24/83 (28%), Positives = 39/83 (46%), Gaps = 7/83 (8%)
Query 5 SRLSKMSSDAQVAPSRMTLQVAKQKKKG-----ASQGYQLLK--KKSDALSARFRGMLKE 57
+R + + A+ AP R+ V + K+ A+ +L SDA + RFRG L E
Sbjct 825 ARAATTCAGARTAPQRLEATVEGKAKRQLVSSCAASSLSVLAGGHASDAAACRFRGCLAE 884
Query 58 IVKTKLSIGDTINEAHFSMAKAS 80
+++ + + EAH S A S
Sbjct 885 RKQSRRELRREVPEAHASKASCS 907
> cel:R06C1.1 hda-3; Histone DeAcetylase family member (hda-3);
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=465
Score = 33.1 bits (74), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 32/118 (27%), Positives = 58/118 (49%), Gaps = 14/118 (11%)
Query 90 QLMQRIKRPAVFVTAAYDNVAGVRLPVFQITT---DPTVDILKNIN-----LSAGGHVI- 140
++M R + AV + D++AG RL VF +TT V+ +K+ N + GG+ I
Sbjct 246 EVMARFQPEAVVLQCGADSLAGDRLGVFNLTTYGHGKCVEYMKSFNVPLLLVGGGGYTIR 305
Query 141 -LAARDKYQEALA---ELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPK-LDKSI 193
++ Y+ A+A E+ L F + K+ + + AL+N P+ +D++I
Sbjct 306 NVSRCWLYETAIALNQEVSDDLPLHDYFDYFIPDYKLHIKPLAALSNFNTPEFIDQTI 363
> tgo:TGME49_015080 hypothetical protein
Length=3347
Score = 30.8 bits (68), Expect = 3.1, Method: Composition-based stats.
Identities = 32/131 (24%), Positives = 54/131 (41%), Gaps = 5/131 (3%)
Query 82 AGGNDLREQLMQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVIL 141
AGG Q Q ++ P+ F + + RL + T D L++ S +
Sbjct 2868 AGGLPASLQQAQHLRLPSRFRSTLRSRL---RLLPGRRTARVREDALESRERSDASEEMA 2924
Query 142 AARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELD 201
D + LA + +A+ Q + F L ++ L N V PK+DK + + +
Sbjct 2925 TVNDAEKRRLASQLLVATGQKSLFLLRPRFELVEETEGLLQNEVGPKMDKEVEQVLETNQ 2984
Query 202 EME--REEFSG 210
E E R+E +G
Sbjct 2985 EKESLRKETTG 2995
> pfa:PF07_0086 conserved Plasmodium membrane protein, unknown
function
Length=3429
Score = 30.8 bits (68), Expect = 3.3, Method: Composition-based stats.
Identities = 13/40 (32%), Positives = 25/40 (62%), Gaps = 1/40 (2%)
Query 167 LDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMERE 206
++ E+ N VN +N V K+D+ +N + KE+++M +E
Sbjct 795 MNEEVNKMNEEVNKMNKEV-NKMDEEVNKMNKEVNKMNKE 833
Lambda K H
0.317 0.130 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6559193640
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40