bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3700_orf1
Length=102
Score E
Sequences producing significant alignments: (Bits) Value
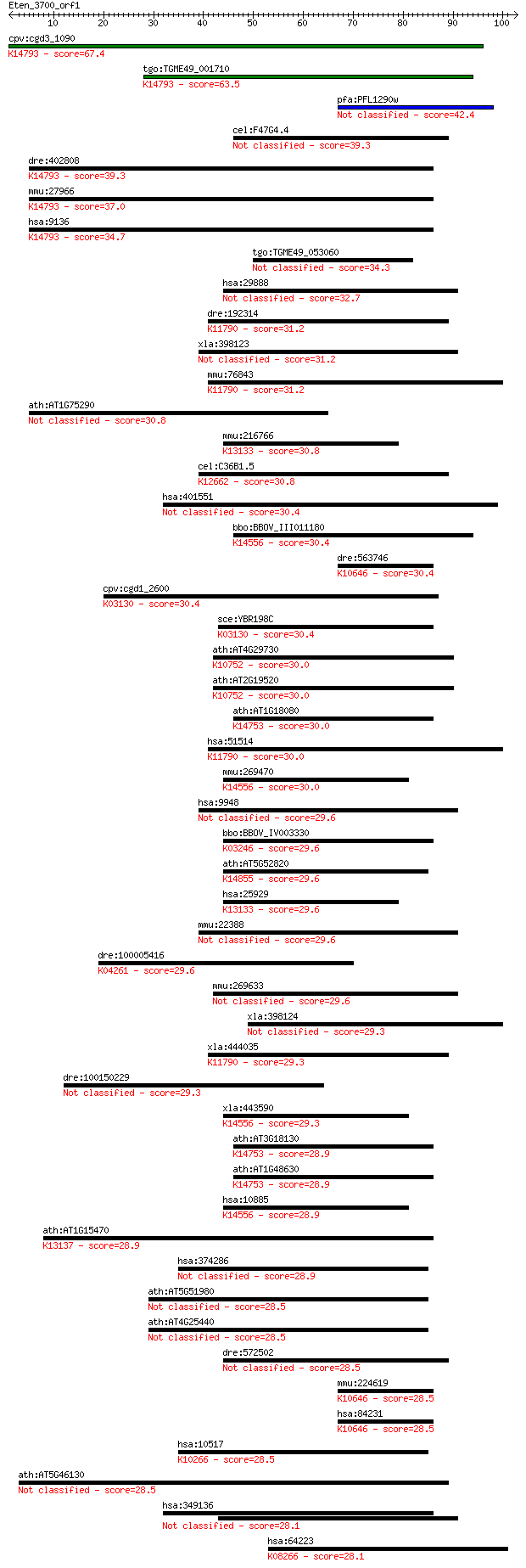
cpv:cgd3_1090 Rrp9p/U3-55K-family snoRNP-associated protein wi... 67.4 1e-11
tgo:TGME49_001710 U3 small nucleolar ribonucleoprotein complex... 63.5 2e-10
pfa:PFL1290w hypothetical protein 42.4 4e-04
cel:F47G4.4 hypothetical protein 39.3 0.003
dre:402808 rrp9, rnu3ip2; ribosomal RNA processing 9, small su... 39.3 0.003
mmu:27966 Rrp9, 55kDa, D19435, D9Wsu10e, MGC25949, Rnu3ip2, U3... 37.0 0.017
hsa:9136 RRP9, RNU3IP2, U3-55K; ribosomal RNA processing 9, sm... 34.7 0.084
tgo:TGME49_053060 hypothetical protein 34.3 0.097
hsa:29888 STRN4, FLJ35594, ZIN, zinedin; striatin, calmodulin ... 32.7 0.25
dre:192314 dtl, cb151, chunp6871, wu:fb54b02; denticleless hom... 31.2 0.84
xla:398123 wdr1-a, MGC52751, aip1, wdr1b; WD repeat domain 1 31.2
mmu:76843 Dtl, 2810047L02Rik, 5730564G15Rik, Ramp; denticleles... 31.2 0.93
ath:AT1G75290 oxidoreductase, acting on NADH or NADPH 30.8 1.1
mmu:216766 Gemin5, AA407055, AA407208, AI451603, BB194447, C33... 30.8 1.1
cel:C36B1.5 prp-4; yeast PRP (splicing factor) related family ... 30.8 1.2
hsa:401551 WDR38; WD repeat domain 38 30.4
bbo:BBOV_III011180 17.m07962; WD domain, G-beta repeat contain... 30.4 1.4
dre:563746 traf7, MGC158391, im:7149067, zgc:158391; TNF recep... 30.4 1.5
cpv:cgd1_2600 transcription factor TAF5p, TBP associated prote... 30.4 1.5
sce:YBR198C TAF5, TAF90; Taf5p; K03130 transcription initiatio... 30.4 1.6
ath:AT4G29730 NFC5; NFC5 (Nucleosome/chromatin assembly factor... 30.0 1.6
ath:AT2G19520 FVE; FVE; metal ion binding; K10752 histone-bind... 30.0 1.7
ath:AT1G18080 ATARCA; ATARCA; nucleotide binding; K14753 guani... 30.0 1.8
hsa:51514 DTL, CDT2, DCAF2, L2DTL, RAMP; denticleless homolog ... 30.0 2.0
mmu:269470 Wdr3, AW546279, D030020G18Rik; WD repeat domain 3; ... 30.0 2.1
hsa:9948 WDR1, AIP1, NORI-1; WD repeat domain 1 29.6
bbo:BBOV_IV003330 21.m02790; translation initiation factor 3 s... 29.6 2.2
ath:AT5G52820 WD-40 repeat family protein / notchless protein,... 29.6 2.3
hsa:25929 GEMIN5, DKFZp586M1824, MGC142174; gem (nuclear organ... 29.6 2.4
mmu:22388 Wdr1, Aip1, D5Wsu185e, rede; WD repeat domain 1 29.6
dre:100005416 ptger4b, si:dkey-204l11.6; prostaglandin E recep... 29.6 2.6
mmu:269633 Wdr86, 2810046M22Rik, BC059069; WD repeat domain 86 29.6
xla:398124 frl2; FRL2 protein 29.3 3.0
xla:444035 dtl-b, MGC82606, cdt2, cdt2-b, dcaf2, dtl, l2dtl, r... 29.3 3.1
dre:100150229 CG3407-like 29.3 3.3
xla:443590 wdr3; WD repeat domain 3; K14556 U3 small nucleolar... 29.3 3.4
ath:AT3G18130 RACK1C_AT (RECEPTOR FOR ACTIVATED C KINASE 1 C);... 28.9 3.8
ath:AT1G48630 RACK1B_AT (RECEPTOR FOR ACTIVATED C KINASE 1 B);... 28.9 3.8
hsa:10885 WDR3, FLJ12796; WD repeat domain 3; K14556 U3 small ... 28.9 3.8
ath:AT1G15470 transducin family protein / WD-40 repeat family ... 28.9 3.9
hsa:374286 CDRT1, C170RF1, C17ORF1, C17ORF1A, DKFZp434O1826, H... 28.9 4.6
ath:AT5G51980 WD-40 repeat family protein / zfwd2 protein (ZFW... 28.5 5.0
ath:AT4G25440 ZFWD1; ZFWD1 (zinc finger WD40 repeat protein 1)... 28.5 5.3
dre:572502 WD repeat-containing protein 7-like 28.5 5.3
mmu:224619 Traf7, MGC7807, RFWD1; TNF receptor-associated fact... 28.5 5.8
hsa:84231 TRAF7, DKFZp586I021, MGC7807, RFWD1, RNF119; TNF rec... 28.5 5.8
hsa:10517 FBXW10, Fbw10, HREP, SM25H2, SM2SH2; F-box and WD re... 28.5 5.8
ath:AT5G46130 hypothetical protein 28.5 6.0
hsa:349136 WDR86, FLJ51355, MGC129839; WD repeat domain 86 28.1
hsa:64223 MLST8, GBL, GbetaL, LST8, MGC111011, POP3, WAT1; MTO... 28.1 7.8
> cpv:cgd3_1090 Rrp9p/U3-55K-family snoRNP-associated protein
with several WD40 repeats ; K14793 ribosomal RNA-processing
protein 9
Length=457
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 41/96 (42%), Positives = 52/96 (54%), Gaps = 10/96 (10%)
Query 1 RVDLFDSDTEKEKLVETLKSRKQQQKASFRCVAQGLCL-RDLGFFKGHKLPVTSVALPGL 59
++D D D K+ L + K S R +A + D F+KGHKL T V L
Sbjct 95 KIDALDDDDIKKSL------NILEGKISQRDIADSFNITSDSTFYKGHKLSPTCVTLDKD 148
Query 60 GGPPRCAYTGGKDCCILRWDLQTGQKDIFKGCRNAF 95
G R AYTGGKDC I++WDL+TG+K IF G R F
Sbjct 149 G---RTAYTGGKDCAIIKWDLETGKKIIFPGSRKDF 181
> tgo:TGME49_001710 U3 small nucleolar ribonucleoprotein complex-associated
protein, putative ; K14793 ribosomal RNA-processing
protein 9
Length=555
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 35/87 (40%), Positives = 44/87 (50%), Gaps = 21/87 (24%)
Query 28 SFRCVAQGLCLRD--LGFFKGHKLPVTSVALPGLGGPP-------------------RCA 66
+F +A L L L F +GHK P+T VA PG+G R
Sbjct 150 AFAPIASSLSLSPAPLAFLRGHKRPLTCVAAPGVGDASEATPEPLSSGAAPSASPLDRHI 209
Query 67 YTGGKDCCILRWDLQTGQKDIFKGCRN 93
YTGGKDCC++ WDLQ +K IF+G RN
Sbjct 210 YTGGKDCCVILWDLQEEKKTIFEGARN 236
> pfa:PFL1290w hypothetical protein
Length=636
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 17/32 (53%), Positives = 23/32 (71%), Gaps = 1/32 (3%)
Query 67 YTGGKDCCILRWDLQTGQK-DIFKGCRNAFAA 97
YTGGKD CI+ WD+ G+K I+KG +N+F
Sbjct 325 YTGGKDACIIEWDIIKGEKVHIYKGNKNSFTG 356
> cel:F47G4.4 hypothetical protein
Length=722
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 22/43 (51%), Gaps = 2/43 (4%)
Query 46 GHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQKDIF 88
GH LP+TS+A P C YTG DC +WD + IF
Sbjct 91 GHTLPITSLAASKFN--PYCWYTGSSDCDWTQWDTRMNPSKIF 131
> dre:402808 rrp9, rnu3ip2; ribosomal RNA processing 9, small
subunit (SSU) processome component, homolog (yeast); K14793
ribosomal RNA-processing protein 9
Length=471
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 24/84 (28%), Positives = 46/84 (54%), Gaps = 9/84 (10%)
Query 5 FDSDTEKEKLVETLKSRKQQQKASFRCVAQGLCLRD---LGFFKGHKLPVTSVALPGLGG 61
F++D +L E + +K + + R +A+ L D + +GHKLPVT + +
Sbjct 99 FEADLIAGRLQEDVLEQKGKLQ---RLIAKELKAPDPAEIRLLRGHKLPVTCLVITP--- 152
Query 62 PPRCAYTGGKDCCILRWDLQTGQK 85
+ ++ KDC I++WD+++G+K
Sbjct 153 DEKHIFSASKDCSIIKWDVESGKK 176
> mmu:27966 Rrp9, 55kDa, D19435, D9Wsu10e, MGC25949, Rnu3ip2,
U3-55k; RRP9, small subunit (SSU) processome component, homolog
(yeast); K14793 ribosomal RNA-processing protein 9
Length=475
Score = 37.0 bits (84), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 24/83 (28%), Positives = 44/83 (53%), Gaps = 7/83 (8%)
Query 5 FDSDTEKEKLVE-TLKSRKQQQKASFRCVAQGLCLRDLGFFKGHKLPVTSVALPGLGGPP 63
F+ D +L E L+ R + QK+ + + Q D+ +GH+L +T + + P
Sbjct 103 FEEDQVAGRLKEDVLEQRGRLQKSVAKEI-QAPAPTDIRVLRGHQLSITCLVIT----PD 157
Query 64 RCA-YTGGKDCCILRWDLQTGQK 85
A ++ KDC I++W ++TG+K
Sbjct 158 DLAIFSAAKDCTIIKWSVETGRK 180
> hsa:9136 RRP9, RNU3IP2, U3-55K; ribosomal RNA processing 9,
small subunit (SSU) processome component, homolog (yeast); K14793
ribosomal RNA-processing protein 9
Length=475
Score = 34.7 bits (78), Expect = 0.084, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 43/83 (51%), Gaps = 7/83 (8%)
Query 5 FDSDTEKEKLVE-TLKSRKQQQKASFRCVAQGLCLRDLGFFKGHKLPVTSVALPGLGGPP 63
F+ D +L E L+ R + QK + + Q D+ +GH+L +T + + P
Sbjct 103 FEEDQVAGRLKEDVLEQRGRLQKLVAKEI-QAPASADIRVLRGHQLSITCLVVT----PD 157
Query 64 RCA-YTGGKDCCILRWDLQTGQK 85
A ++ KDC I++W +++G+K
Sbjct 158 DSAIFSAAKDCSIIKWSVESGRK 180
> tgo:TGME49_053060 hypothetical protein
Length=751
Score = 34.3 bits (77), Expect = 0.097, Method: Compositional matrix adjust.
Identities = 17/32 (53%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 50 PVTSVALPGLGGPPRCAYTGGKDCCILRWDLQ 81
PVTSVA+P G C YTGG D + WDL+
Sbjct 292 PVTSVAVPQDGSSSGCLYTGGYDEMLRLWDLR 323
> hsa:29888 STRN4, FLJ35594, ZIN, zinedin; striatin, calmodulin
binding protein 4
Length=760
Score = 32.7 bits (73), Expect = 0.25, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 23/47 (48%), Gaps = 3/47 (6%)
Query 44 FKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQKDIFKG 90
F+ H+ PV +VA+ G Y+GG D CI W + D + G
Sbjct 494 FRAHRGPVLAVAM---GSNSEYCYSGGADACIHSWKIPDLSMDPYDG 537
> dre:192314 dtl, cb151, chunp6871, wu:fb54b02; denticleless homolog
(Drosophila); K11790 denticleless
Length=647
Score = 31.2 bits (69), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 19/48 (39%), Positives = 25/48 (52%), Gaps = 2/48 (4%)
Query 41 LGFFKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQKDIF 88
LG FKGH+ + SVA TGG+D I+ WD + +KD F
Sbjct 132 LGTFKGHQCSLKSVAF--YKQEKAVFSTGGRDGNIMIWDTRCSKKDGF 177
> xla:398123 wdr1-a, MGC52751, aip1, wdr1b; WD repeat domain 1
Length=608
Score = 31.2 bits (69), Expect = 0.84, Method: Composition-based stats.
Identities = 18/52 (34%), Positives = 23/52 (44%), Gaps = 1/52 (1%)
Query 39 RDLGFFKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQKDIFKG 90
R L KGH + + + G YTG D I WD +TG+ D F G
Sbjct 311 RPLRVIKGHNKSIQCMTVNNSDGR-STIYTGSHDGHINYWDAETGENDTFTG 361
> mmu:76843 Dtl, 2810047L02Rik, 5730564G15Rik, Ramp; denticleless
homolog (Drosophila); K11790 denticleless
Length=729
Score = 31.2 bits (69), Expect = 0.93, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 29/59 (49%), Gaps = 2/59 (3%)
Query 41 LGFFKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQKDIFKGCRNAFAASY 99
+G KGH+ + SVA P TGG+D I+ WD + +KD F N + ++
Sbjct 133 MGTCKGHQCSLKSVAFPKFQK--AVFSTGGRDGNIMIWDTRCNKKDGFYRQVNQISGAH 189
> ath:AT1G75290 oxidoreductase, acting on NADH or NADPH
Length=318
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 29/65 (44%), Gaps = 7/65 (10%)
Query 5 FDSDTEKEKLVETLKS---RKQQQKASFRCVAQGLCLRDL--GFFKGHKLPVTSVALPGL 59
F D +K VE KS RK Q + A+G+ L +F G+ LP PGL
Sbjct 114 FGMDVDKSSAVEPAKSAFGRKLQTRRDIE--AEGIPYTYLVTNYFAGYYLPTLVQLEPGL 171
Query 60 GGPPR 64
PPR
Sbjct 172 TSPPR 176
> mmu:216766 Gemin5, AA407055, AA407208, AI451603, BB194447, C330013N08;
gem (nuclear organelle) associated protein 5; K13133
gem associated protein 5
Length=1503
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 19/35 (54%), Gaps = 2/35 (5%)
Query 44 FKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRW 78
F+GH+ + VA + P C Y+G D C+ RW
Sbjct 678 FRGHRGRLLCVAWSPVD--PECIYSGADDFCVYRW 710
> cel:C36B1.5 prp-4; yeast PRP (splicing factor) related family
member (prp-4); K12662 U4/U6 small nuclear ribonucleoprotein
PRP4
Length=496
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 26/50 (52%), Gaps = 3/50 (6%)
Query 39 RDLGFFKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQKDIF 88
++L + +GH V VA G A TGG DC WD++TG+ +F
Sbjct 329 KELLYQEGHSKSVADVAFHPDGS---VALTGGHDCYGRVWDMRTGRCIMF 375
> hsa:401551 WDR38; WD repeat domain 38
Length=314
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 36/68 (52%), Gaps = 7/68 (10%)
Query 32 VAQGLCLRDLGFFKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQK-DIFKG 90
VA+ CLR L KGH+ V +V+ R +GG D ++ WD+Q+GQ + G
Sbjct 92 VARAKCLRVL---KGHQRSVETVSF---SPDSRQLASGGWDKRVMLWDVQSGQMLRLLVG 145
Query 91 CRNAFAAS 98
R++ +S
Sbjct 146 HRDSIQSS 153
> bbo:BBOV_III011180 17.m07962; WD domain, G-beta repeat containing
protein; K14556 U3 small nucleolar RNA-associated protein
12
Length=1005
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 27/50 (54%), Gaps = 6/50 (12%)
Query 46 GHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQKDIFK--GCRN 93
GHK +T +A+ + +G +DCCI+ WD TG +F+ G RN
Sbjct 115 GHKSAITCLAI---SPSQQLLASGSQDCCIIIWD-TTGDLGLFRLEGHRN 160
> dre:563746 traf7, MGC158391, im:7149067, zgc:158391; TNF receptor-associated
factor 7; K10646 E3 ubiquitin-protein ligase
TRAF7 [EC:6.3.2.19]
Length=639
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 11/19 (57%), Positives = 14/19 (73%), Gaps = 0/19 (0%)
Query 67 YTGGKDCCILRWDLQTGQK 85
Y+G DC I+ WD+QT QK
Sbjct 422 YSGSADCTIIVWDIQTLQK 440
> cpv:cgd1_2600 transcription factor TAF5p, TBP associated protein
involved in transcription ; K03130 transcription initiation
factor TFIID subunit 5
Length=753
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 32/68 (47%), Gaps = 8/68 (11%)
Query 20 SRKQQQKASFRCVAQGLCLRDLGFFKGHKLPVTSVAL-PGLGGPPRCAYTGGKDCCILRW 78
S Q A C ++ +R F GH V SV++ P TGG D I+ W
Sbjct 539 SGSSDQSARLWCTSRTYPIR---IFTGHFGDVRSVSIHPN----SSITVTGGSDNQIIIW 591
Query 79 DLQTGQKD 86
D++TG+K+
Sbjct 592 DVRTGKKE 599
> sce:YBR198C TAF5, TAF90; Taf5p; K03130 transcription initiation
factor TFIID subunit 5
Length=798
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 23/43 (53%), Gaps = 3/43 (6%)
Query 43 FFKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQK 85
F GH PV S+A+ G R TG +D I WD+ TG++
Sbjct 646 LFLGHTAPVISIAVCPDG---RWLSTGSEDGIINVWDIGTGKR 685
> ath:AT4G29730 NFC5; NFC5 (Nucleosome/chromatin assembly factor
group C 5); K10752 histone-binding protein RBBP4
Length=487
Score = 30.0 bits (66), Expect = 1.6, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 21/48 (43%), Gaps = 2/48 (4%)
Query 42 GFFKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQKDIFK 89
G + GHK V VA C+ G D C++ WD +TG K
Sbjct 266 GIYNGHKDTVEDVAFCPSSAQEFCSV--GDDSCLMLWDARTGTSPAMK 311
> ath:AT2G19520 FVE; FVE; metal ion binding; K10752 histone-binding
protein RBBP4
Length=507
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 22/48 (45%), Gaps = 2/48 (4%)
Query 42 GFFKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQKDIFK 89
G + GH+ V VA C+ G D C++ WD +TG + K
Sbjct 286 GVYHGHEDTVEDVAFSPTSAQEFCSV--GDDSCLILWDARTGTNPVTK 331
> ath:AT1G18080 ATARCA; ATARCA; nucleotide binding; K14753 guanine
nucleotide-binding protein subunit beta-2-like 1 protein
Length=327
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 18/40 (45%), Positives = 24/40 (60%), Gaps = 3/40 (7%)
Query 46 GHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQK 85
GH V++VA+ G CA +GGKD +L WDL G+K
Sbjct 192 GHTGYVSTVAVSPDGS--LCA-SGGKDGVVLLWDLAEGKK 228
> hsa:51514 DTL, CDT2, DCAF2, L2DTL, RAMP; denticleless homolog
(Drosophila); K11790 denticleless
Length=730
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 29/59 (49%), Gaps = 2/59 (3%)
Query 41 LGFFKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQKDIFKGCRNAFAASY 99
+G KGH+ + SVA C TGG+D I+ WD + +KD F N + ++
Sbjct 133 IGTCKGHQCSLKSVAFSKFEKAVFC--TGGRDGNIMVWDTRCNKKDGFYRQVNQISGAH 189
> mmu:269470 Wdr3, AW546279, D030020G18Rik; WD repeat domain 3;
K14556 U3 small nucleolar RNA-associated protein 12
Length=942
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 17/37 (45%), Positives = 22/37 (59%), Gaps = 3/37 (8%)
Query 44 FKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDL 80
F GHK VTS+ LGG R A +G KD ++ WD+
Sbjct 103 FNGHKAAVTSLKYDQLGG--RLA-SGSKDTDVIIWDV 136
> hsa:9948 WDR1, AIP1, NORI-1; WD repeat domain 1
Length=466
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 24/52 (46%), Gaps = 1/52 (1%)
Query 39 RDLGFFKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQKDIFKG 90
+ L KGH + + + GG Y+G D I WD +TG+ D F G
Sbjct 171 KPLHVIKGHSKSIQCLTVHKNGGKSYI-YSGSHDGHINYWDSETGENDSFAG 221
> bbo:BBOV_IV003330 21.m02790; translation initiation factor 3
subunit 2; K03246 translation initiation factor 3 subunit I
Length=340
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 21/42 (50%), Gaps = 3/42 (7%)
Query 44 FKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQK 85
+GH+ P+T V G +T GKD C++ W GQ+
Sbjct 6 LRGHRRPLTCVKTNREGD---LLFTCGKDACLMLWRTDNGQQ 44
> ath:AT5G52820 WD-40 repeat family protein / notchless protein,
putative; K14855 ribosome assembly protein 4
Length=473
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 19/41 (46%), Gaps = 4/41 (9%)
Query 44 FKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQ 84
GH L VT V G G YTG +DC I W+ G+
Sbjct 237 LSGHTLAVTCVKWGGDG----IIYTGSQDCTIKMWETTQGK 273
> hsa:25929 GEMIN5, DKFZp586M1824, MGC142174; gem (nuclear organelle)
associated protein 5; K13133 gem associated protein
5
Length=1508
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 19/35 (54%), Gaps = 2/35 (5%)
Query 44 FKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRW 78
F+GH+ + VA L P C Y+G D C+ +W
Sbjct 678 FRGHRGRLLCVAWSPLD--PDCIYSGADDFCVHKW 710
> mmu:22388 Wdr1, Aip1, D5Wsu185e, rede; WD repeat domain 1
Length=606
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 24/52 (46%), Gaps = 1/52 (1%)
Query 39 RDLGFFKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQKDIFKG 90
+ L KGH + + + GG Y+G D I WD +TG+ D F G
Sbjct 311 KPLRVIKGHSKSIQCLTVHRNGGKSYI-YSGSHDGHINYWDSETGENDSFSG 361
> dre:100005416 ptger4b, si:dkey-204l11.6; prostaglandin E receptor
4 (subtype EP4) b; K04261 prostaglandin E receptor 4
Length=497
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 19/53 (35%), Positives = 28/53 (52%), Gaps = 2/53 (3%)
Query 19 KSRKQQQKASFRCVAQGLCLRD-LGFFKGHKLPV-TSVALPGLGGPPRCAYTG 69
KSRK+Q++ +F + GL + D LG + + T V GG P C Y+G
Sbjct 72 KSRKEQKETTFYTLVCGLAVTDLLGTLLASPVTIATYVKGEWPGGMPLCQYSG 124
> mmu:269633 Wdr86, 2810046M22Rik, BC059069; WD repeat domain
86
Length=380
Score = 29.6 bits (65), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 25/50 (50%), Gaps = 6/50 (12%)
Query 42 GFFKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQ-KDIFKG 90
+GH+ VT L A+T DC I RWD++TGQ +++G
Sbjct 51 ALLQGHESYVTFCHLE-----DEAAFTCSADCTIRRWDVRTGQCLQVYRG 95
> xla:398124 frl2; FRL2 protein
Length=169
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 16/52 (30%), Positives = 24/52 (46%), Gaps = 1/52 (1%)
Query 49 LPVTSVALPG-LGGPPRCAYTGGKDCCILRWDLQTGQKDIFKGCRNAFAASY 99
LP+T L G PP CAY + ++ + F G +++F ASY
Sbjct 96 LPLTDCLLMGRTARPPNCAYNQTRTTGVINITCENNYPVHFAGYKSSFCASY 147
> xla:444035 dtl-b, MGC82606, cdt2, cdt2-b, dcaf2, dtl, l2dtl,
ramp; denticleless homolog; K11790 denticleless
Length=711
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 26/49 (53%), Gaps = 4/49 (8%)
Query 41 LGFFKGHKLPVTSVALPGLGGPPRCAY-TGGKDCCILRWDLQTGQKDIF 88
+G KGH+ + SVA R + TGG+D I+ WD + +KD F
Sbjct 133 IGECKGHQCSLKSVAFSKF---ERAVFSTGGRDGNIMVWDTRCNKKDGF 178
> dre:100150229 CG3407-like
Length=466
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 26/64 (40%), Gaps = 12/64 (18%)
Query 12 EKLVETLKSRKQQQKA-----SFRCVAQGLCLRDLGFFKGHKL-------PVTSVALPGL 59
EK L S + Q+ +RC G C R+LG K H+L PV + P L
Sbjct 388 EKRFRHLDSLHKHQRIHTGERPYRCAECGCCFRELGQLKKHRLKHSPTFVPVANPTFPLL 447
Query 60 GGPP 63
P
Sbjct 448 PAGP 451
> xla:443590 wdr3; WD repeat domain 3; K14556 U3 small nucleolar
RNA-associated protein 12
Length=942
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 20/37 (54%), Gaps = 3/37 (8%)
Query 44 FKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDL 80
F GHK VT++ LGG +G KD ++ WD+
Sbjct 103 FNGHKAAVTTLNYDHLGGR---LVSGSKDTDVIVWDV 136
> ath:AT3G18130 RACK1C_AT (RECEPTOR FOR ACTIVATED C KINASE 1 C);
nucleotide binding; K14753 guanine nucleotide-binding protein
subunit beta-2-like 1 protein
Length=326
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 18/40 (45%), Positives = 23/40 (57%), Gaps = 3/40 (7%)
Query 46 GHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQK 85
GH + +VA+ G CA +GGKD IL WDL G+K
Sbjct 191 GHSGYLNTVAVSPDGS--LCA-SGGKDGVILLWDLAEGKK 227
> ath:AT1G48630 RACK1B_AT (RECEPTOR FOR ACTIVATED C KINASE 1 B);
nucleotide binding; K14753 guanine nucleotide-binding protein
subunit beta-2-like 1 protein
Length=326
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 18/40 (45%), Positives = 23/40 (57%), Gaps = 3/40 (7%)
Query 46 GHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQK 85
GH + +VA+ G CA +GGKD IL WDL G+K
Sbjct 191 GHSGYLNTVAVSPDGS--LCA-SGGKDGVILLWDLAEGKK 227
> hsa:10885 WDR3, FLJ12796; WD repeat domain 3; K14556 U3 small
nucleolar RNA-associated protein 12
Length=943
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 22/37 (59%), Gaps = 3/37 (8%)
Query 44 FKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDL 80
F GHK +T++ LGG R A +G KD I+ WD+
Sbjct 103 FNGHKAAITTLKYDQLGG--RLA-SGSKDTDIIVWDV 136
> ath:AT1G15470 transducin family protein / WD-40 repeat family
protein; K13137 serine-threonine kinase receptor-associated
protein
Length=333
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 24/91 (26%), Positives = 42/91 (46%), Gaps = 15/91 (16%)
Query 8 DTEKEKLVETLKSR------KQQQKASFRCVAQGLCLR-----DLGFFKGHKLP--VTSV 54
D +K+V TL+++ + Q + A G ++ + G K + +P V S
Sbjct 169 DIRSDKIVHTLETKSPVTSAEVSQDGRYITTADGSSVKFWDAKNFGLLKSYDMPCNVESA 228
Query 55 ALPGLGGPPRCAYTGGKDCCILRWDLQTGQK 85
+L G A GG+D + R+D QTG++
Sbjct 229 SLEPKHGNTFIA--GGEDMWVHRFDFQTGEE 257
> hsa:374286 CDRT1, C170RF1, C17ORF1, C17ORF1A, DKFZp434O1826,
HREP, SM25H2; CMT1A duplicated region transcript 1
Length=752
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 26/53 (49%), Gaps = 14/53 (26%)
Query 35 GLCLRDLGFFKGHKLPVTSVALPGLGGPPRCA---YTGGKDCCILRWDLQTGQ 84
G+C R F GH+ +T + L C +GG+DC + WD+ TG+
Sbjct 485 GVCTR---IFGGHQGTITCMDL--------CKNRLVSGGRDCQVKVWDVDTGK 526
> ath:AT5G51980 WD-40 repeat family protein / zfwd2 protein (ZFWD2),
putative
Length=443
Score = 28.5 bits (62), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 27/56 (48%), Gaps = 3/56 (5%)
Query 29 FRCVAQGLCLRDLGFFKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQ 84
C ++G L GH+ V+ +ALP G + YTG KD + WD +GQ
Sbjct 132 LHCWSKGESFALLTQLDGHEKLVSGIALPS--GSDKL-YTGSKDETLRVWDCASGQ 184
> ath:AT4G25440 ZFWD1; ZFWD1 (zinc finger WD40 repeat protein
1); nucleic acid binding / zinc ion binding
Length=430
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 26/56 (46%), Gaps = 3/56 (5%)
Query 29 FRCVAQGLCLRDLGFFKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQ 84
C ++G L GH+ VT +ALP G + YT KD + WD +GQ
Sbjct 125 LHCWSKGDSFSLLTQLDGHQKVVTGIALPS--GSDKL-YTASKDETVRIWDCASGQ 177
> dre:572502 WD repeat-containing protein 7-like
Length=1059
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 25/49 (51%), Gaps = 4/49 (8%)
Query 44 FKGHKLPVTSVALPGLGGP---PRCAYTGGKDCCILRWDLQTGQ-KDIF 88
+GH+ VT + P P R +GG D ++ WD+ TG+ K IF
Sbjct 463 LRGHRHKVTCLLYPHQVSPRYDQRSLVSGGVDFSVIIWDIFTGEMKHIF 511
> mmu:224619 Traf7, MGC7807, RFWD1; TNF receptor-associated factor
7; K10646 E3 ubiquitin-protein ligase TRAF7 [EC:6.3.2.19]
Length=669
Score = 28.5 bits (62), Expect = 5.8, Method: Composition-based stats.
Identities = 10/19 (52%), Positives = 13/19 (68%), Gaps = 0/19 (0%)
Query 67 YTGGKDCCILRWDLQTGQK 85
Y+G DC I+ WD+Q QK
Sbjct 452 YSGSADCTIIVWDIQNLQK 470
> hsa:84231 TRAF7, DKFZp586I021, MGC7807, RFWD1, RNF119; TNF receptor-associated
factor 7; K10646 E3 ubiquitin-protein ligase
TRAF7 [EC:6.3.2.19]
Length=670
Score = 28.5 bits (62), Expect = 5.8, Method: Composition-based stats.
Identities = 10/19 (52%), Positives = 13/19 (68%), Gaps = 0/19 (0%)
Query 67 YTGGKDCCILRWDLQTGQK 85
Y+G DC I+ WD+Q QK
Sbjct 453 YSGSADCTIIVWDIQNLQK 471
> hsa:10517 FBXW10, Fbw10, HREP, SM25H2, SM2SH2; F-box and WD
repeat domain containing 10; K10266 F-box and WD-40 domain protein
10
Length=1051
Score = 28.5 bits (62), Expect = 5.8, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 26/53 (49%), Gaps = 14/53 (26%)
Query 35 GLCLRDLGFFKGHKLPVTSVALPGLGGPPRCA---YTGGKDCCILRWDLQTGQ 84
G+C R F GH+ +T + L C +GG+DC + WD+ TG+
Sbjct 485 GVCTR---IFGGHQGTITCMDL--------CKNRLVSGGRDCQVKVWDVDTGK 526
> ath:AT5G46130 hypothetical protein
Length=376
Score = 28.5 bits (62), Expect = 6.0, Method: Composition-based stats.
Identities = 22/86 (25%), Positives = 36/86 (41%), Gaps = 2/86 (2%)
Query 3 DLFDSDTEKEKLVETLKSRKQQQKASFRCVAQGLCLRDLGFFKGHKLPVTSVALPGLGGP 62
D F+SDT K L ++++ + + + DL F GH P+ A G
Sbjct 269 DAFESDTVVHKTKRFLVFKEKENAKRHKDMYYTENIGDLCIFLGHGEPMCVPASSSPGLK 328
Query 63 PRCAYTGGKDCCILRWDLQTGQKDIF 88
P C Y G + + +D+ T +F
Sbjct 329 PNCIYFAGHNFGV--YDITTQTITLF 352
> hsa:349136 WDR86, FLJ51355, MGC129839; WD repeat domain 86
Length=376
Score = 28.1 bits (61), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 24/49 (48%), Gaps = 6/49 (12%)
Query 43 FFKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQ-KDIFKG 90
+GH+ VT L A+T DC I RWD+ TGQ +++G
Sbjct 52 LLQGHESYVTFCQLE-----DEAAFTCSADCTIRRWDVLTGQCLQVYRG 95
Score = 28.1 bits (61), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 26/54 (48%), Gaps = 6/54 (11%)
Query 32 VAQGLCLRDLGFFKGHKLPVTSVALPGLGGPPRCAYTGGKDCCILRWDLQTGQK 85
VA G C + L +GH V + L P A+TG D I WD+ +G++
Sbjct 180 VASGCCHQTL---RGHTGAVLCLVLDT---PGHTAFTGSTDATIRAWDILSGEQ 227
> hsa:64223 MLST8, GBL, GbetaL, LST8, MGC111011, POP3, WAT1; MTOR
associated protein, LST8 homolog (S. cerevisiae); K08266
G protein beta subunit-like
Length=326
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 22/48 (45%), Gaps = 4/48 (8%)
Query 53 SVALPGLGGPPRCAYTGGKDCCILRWDLQTGQKDIFKGCRNAFAASYP 100
++A G R YTGG+DC WDL++ C+ F + P
Sbjct 87 NIASVGFHEDGRWMYTGGEDCTARIWDLRSRNLQ----CQRIFQVNAP 130
Lambda K H
0.324 0.141 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2031832220
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40