bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3783_orf2
Length=209
Score E
Sequences producing significant alignments: (Bits) Value
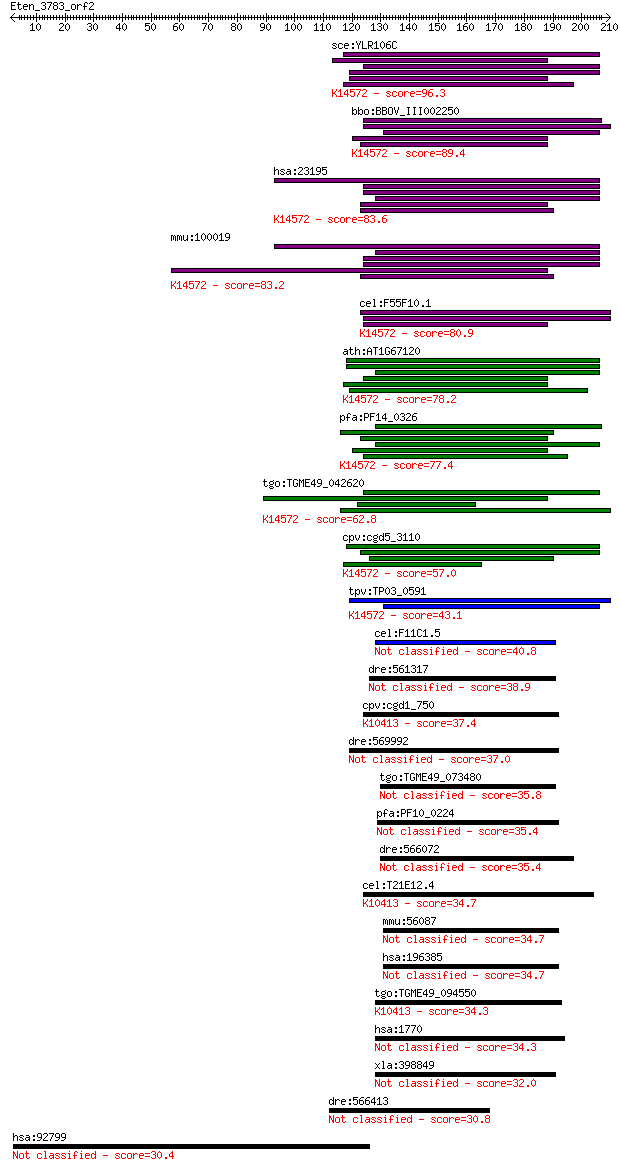
sce:YLR106C MDN1, REA1; Huge dynein-related AAA-type ATPase (m... 96.3 7e-20
bbo:BBOV_III002250 17.m07217; ATPase associated with various c... 89.4 8e-18
hsa:23195 MDN1, DKFZp686H16106, FLJ23395, FLJ25587, FLJ42031, ... 83.6 4e-16
mmu:100019 Mdn1, 4833432B22Rik, A130070M06, AA958993, D4Abb1e,... 83.2 6e-16
cel:F55F10.1 hypothetical protein; K14572 midasin 80.9 3e-15
ath:AT1G67120 ATP binding / ATPase/ nucleoside-triphosphatase/... 78.2 2e-14
pfa:PF14_0326 dynein-related AAA-type ATPase; K14572 midasin 77.4 3e-14
tgo:TGME49_042620 hypothetical protein ; K14572 midasin 62.8 9e-10
cpv:cgd5_3110 MDN1, midasin ; K14572 midasin 57.0 5e-08
tpv:TP03_0591 hypothetical protein; K14572 midasin 43.1 6e-04
cel:F11C1.5 hypothetical protein 40.8 0.003
dre:561317 si:dkey-18l1.1 38.9 0.012
cpv:cgd1_750 dynein heavy chain ; K10413 dynein heavy chain 1,... 37.4 0.037
dre:569992 dynein, axonemal, heavy chain 5-like 37.0 0.050
tgo:TGME49_073480 axonemal beta dynein heavy chain, putative (... 35.8 0.12
pfa:PF10_0224 dynein heavy chain, putative; K06025 [EC:3.6.4.2] 35.4 0.14
dre:566072 dynein, axonemal, heavy polypeptide 11-like 35.4 0.16
cel:T21E12.4 dhc-1; Dynein Heavy Chain family member (dhc-1); ... 34.7 0.23
mmu:56087 Dnahc10; dynein, axonemal, heavy chain 10 34.7
hsa:196385 DNAH10, FLJ38262, FLJ43486, FLJ43808, KIAA2017; dyn... 34.7 0.26
tgo:TGME49_094550 dynein heavy chain, putative ; K10413 dynein... 34.3 0.30
hsa:1770 DNAH9, DNAH17L, DNEL1, DYH9, Dnahc9, HL-20, HL20, KIA... 34.3 0.33
xla:398849 MGC68485; KIAA0564 32.0 1.6
dre:566413 si:dkey-20n10.1 30.8 3.2
hsa:92799 SHKBP1, Sb1; SH3KBP1 binding protein 1 30.4
> sce:YLR106C MDN1, REA1; Huge dynein-related AAA-type ATPase
(midasin), forms extended pre-60S particle with the Rix1 complex
(Rix1p-Ipi1p-Ipi3p), may mediate ATP-dependent remodeling
of 60S subunits and subsequent export from nucleoplasm to
cytoplasm; K14572 midasin
Length=4910
Score = 96.3 bits (238), Expect = 7e-20, Method: Composition-based stats.
Identities = 50/93 (53%), Positives = 62/93 (66%), Gaps = 5/93 (5%)
Query 117 SQEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLT----EG 172
++E +FEW D LV AV KG WL+L +A+LCS SVLDRLN+LLE GSLL+ E
Sbjct 2185 TKEASVKFEWFDGMLVKAVEKGHWLILDNANLCSPSVLDRLNSLLEIDGSLLINECSQED 2244
Query 173 GAPREVKRHPNFRILLTAEAANTHLLSAALRNR 205
G PR +K HPNFR+ LT + LS A+RNR
Sbjct 2245 GQPRVLKPHPNFRLFLTMDPKYGE-LSRAMRNR 2276
Score = 58.5 bits (140), Expect = 1e-08, Method: Composition-based stats.
Identities = 30/77 (38%), Positives = 46/77 (59%), Gaps = 2/77 (2%)
Query 113 AAACSQEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLE--DGGSLLLT 170
A + S E F +V+ +LV +R G+WLLL + +L +A L+ ++ LL D S+LL+
Sbjct 798 AQSSSIENSFVFNFVEGSLVKTIRAGEWLLLDEVNLATADTLESISDLLTEPDSRSILLS 857
Query 171 EGGAPREVKRHPNFRIL 187
E G +K HP+FRI
Sbjct 858 EKGDAEPIKAHPDFRIF 874
Score = 58.2 bits (139), Expect = 2e-08, Method: Composition-based stats.
Identities = 36/86 (41%), Positives = 46/86 (53%), Gaps = 4/86 (4%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPRE-VKRHP 182
FEW D L+ A+R G + LL + L SVL+RLN++LE SLLL E G+ V
Sbjct 1472 FEWSDGPLIQAMRTGNFFLLDEISLADDSVLERLNSVLEPERSLLLAEQGSSDSLVTASE 1531
Query 183 NFRILLTAEAANTH---LLSAALRNR 205
NF+ T + LS ALRNR
Sbjct 1532 NFQFFATMNPGGDYGKKELSPALRNR 1557
Score = 49.7 bits (117), Expect = 7e-06, Method: Composition-based stats.
Identities = 29/91 (31%), Positives = 46/91 (50%), Gaps = 6/91 (6%)
Query 119 EEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREV 178
++ + + + LV A+RKG W++L + +L VL+ LN LL+D L + E V
Sbjct 1125 DDTGKLSFKEGVLVEALRKGYWIVLDELNLAPTDVLEALNRLLDDNRELFIPE--TQEVV 1182
Query 179 KRHPNFRILLTAEAANTH----LLSAALRNR 205
HP+F + T + +LS A RNR
Sbjct 1183 HPHPDFLLFATQNPPGIYGGRKILSRAFRNR 1213
Score = 47.8 bits (112), Expect = 2e-05, Method: Composition-based stats.
Identities = 25/69 (36%), Positives = 38/69 (55%), Gaps = 2/69 (2%)
Query 119 EEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREV 178
E +F W D+ + A++KG+W+LL + +L S SVL+ LNA L+ G + E
Sbjct 1790 ERSGEFLWHDAPFLRAMKKGEWVLLDEMNLASQSVLEGLNACLDHRGEAYIPE--LDISF 1847
Query 179 KRHPNFRIL 187
HPNF +
Sbjct 1848 SCHPNFLVF 1856
Score = 35.8 bits (81), Expect = 0.12, Method: Composition-based stats.
Identities = 23/80 (28%), Positives = 36/80 (45%), Gaps = 3/80 (3%)
Query 117 SQEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPR 176
S ++ FEW L AV++G+W+L+ D VL L +LLE + + G
Sbjct 358 SGDKPGTFEWRAGVLATAVKEGRWVLIEDIDKAPTDVLSILLSLLEKRELTIPSRG---E 414
Query 177 EVKRHPNFRILLTAEAANTH 196
VK F+++ T H
Sbjct 415 TVKAANGFQLISTVRINEDH 434
> bbo:BBOV_III002250 17.m07217; ATPase associated with various
cellular activities (AAA) family protein; K14572 midasin
Length=4334
Score = 89.4 bits (220), Expect = 8e-18, Method: Composition-based stats.
Identities = 42/84 (50%), Positives = 56/84 (66%), Gaps = 1/84 (1%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGG-APREVKRHP 182
F WVDSALV ++ G W+LL H + ++LDRLN+LLE GGS L E G R ++ HP
Sbjct 1774 FRWVDSALVRSIECGTWVLLSGIHNTNPAILDRLNSLLESGGSFSLNESGDCNRVIRPHP 1833
Query 183 NFRILLTAEAANTHLLSAALRNRC 206
NFRI +TA+++ +S A RNRC
Sbjct 1834 NFRIFMTADSSYVGKISRAFRNRC 1857
Score = 59.3 bits (142), Expect = 9e-09, Method: Composition-based stats.
Identities = 33/91 (36%), Positives = 48/91 (52%), Gaps = 5/91 (5%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAP--REVKRH 181
FEWVD L ++ KG W L + L +VL+++N++LE +L + E G R +K H
Sbjct 1180 FEWVDGPLTRSMEKGHWFLADEISLADDAVLEKMNSVLESESTLTIPEAGGRNLRVLKAH 1239
Query 182 PNFRILLTAEAANTH---LLSAALRNRCCLV 209
FR L T + H LS AL NR ++
Sbjct 1240 EQFRFLATMNPSGDHGKRELSPALLNRFTVI 1270
Score = 47.0 bits (110), Expect = 4e-05, Method: Composition-based stats.
Identities = 26/80 (32%), Positives = 43/80 (53%), Gaps = 7/80 (8%)
Query 131 LVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRILLTA 190
+V A+++G W++L + +L + VL+ +N +L+D + + E G +K HP F I T
Sbjct 788 IVTAMKQGDWVILDELNLAPSQVLEAINRILDDNREIYIPETG--ETIKSHPEFMIFATQ 845
Query 191 EAANT-----HLLSAALRNR 205
AN+ LS A NR
Sbjct 846 NPANSIYGGRKQLSKAFCNR 865
Score = 39.3 bits (90), Expect = 0.009, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 35/68 (51%), Gaps = 2/68 (2%)
Query 120 EMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVK 179
+ S+F W + ++ A G W +L + +L S VL+ LNALL+ + E R +K
Sbjct 1479 DASRFRWHNGPVLDAAINGHWAVLDELNLASQQVLEGLNALLDHRRETFIPE--LDRFIK 1536
Query 180 RHPNFRIL 187
H FR+
Sbjct 1537 CHDKFRLF 1544
Score = 37.7 bits (86), Expect = 0.027, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 38/66 (57%), Gaps = 2/66 (3%)
Query 123 QFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLED-GGSLLLTEGGAPREVKRH 181
QF++ D L+ A+++G W+LL + +L + +L R +L + + L E G+ R + H
Sbjct 458 QFKFEDGLLIRAMQEGWWILLDEINLAPSDLLQRFVGILSNRRRTFELYECGS-RVIDIH 516
Query 182 PNFRIL 187
NFRI
Sbjct 517 ENFRIF 522
> hsa:23195 MDN1, DKFZp686H16106, FLJ23395, FLJ25587, FLJ42031,
FLJ43191, KIAA0301; MDN1, midasin homolog (yeast); K14572
midasin
Length=5596
Score = 83.6 bits (205), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 46/117 (39%), Positives = 66/117 (56%), Gaps = 8/117 (6%)
Query 93 AGLAHVLESVSSSSAAAAAAAAACSQEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSAS 152
A A ++E S A+ S FEWVDS LV A++ G WLL+ + + C+ S
Sbjct 2194 AEFAKLVEEFRSFGVKLTQLASGHSH---GTFEWVDSMLVQALKSGDWLLMDNVNFCNPS 2250
Query 153 VLDRLNALLEDGGSLLLTE----GGAPREVKRHPNFRILLTAEAANTHLLSAALRNR 205
VLDRLNALLE GG L ++E G+ + +PNFR+ L+ + + +S A+RNR
Sbjct 2251 VLDRLNALLEPGGVLTISERGMIDGSTPTITPNPNFRLFLSMDPVHGD-ISRAMRNR 2306
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 31/86 (36%), Positives = 49/86 (56%), Gaps = 4/86 (4%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLE-DGGSLLLTEGGAPREVKRHP 182
F +V+ L AV+KG+W+LL + +L + +L+ L+ LLE GSL+L + G + RHP
Sbjct 816 FAFVEGTLAQAVKKGEWILLDEINLAAPEILECLSGLLEGSSGSLVLLDRGDTEPLVRHP 875
Query 183 NFRI---LLTAEAANTHLLSAALRNR 205
+FR+ + A L +RNR
Sbjct 876 DFRLFACMNPATDVGKRNLPPGIRNR 901
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 47/92 (51%), Gaps = 10/92 (10%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRH-- 181
FEW D LV A+++ + LL + L SVL+RLN++LE SL+L E G+P +
Sbjct 1448 FEWHDGPLVQAMKEDGFFLLDEISLADDSVLERLNSVLEVEKSLVLAEKGSPEDKDSEIE 1507
Query 182 -----PNFRILLTAEAA---NTHLLSAALRNR 205
FRIL T LS ALRNR
Sbjct 1508 LLTAGKKFRILATMNPGGDFGKKELSPALRNR 1539
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 31/82 (37%), Positives = 44/82 (53%), Gaps = 6/82 (7%)
Query 128 DSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRIL 187
+ L+ A+RKG W++L + +L VL+ LN LL+D LL+TE VK HP F +
Sbjct 1135 EGVLIDAMRKGYWIILDELNLAPTDVLEALNRLLDDNRELLVTE--TQEVVKAHPRFMLF 1192
Query 188 LTAEAANTH----LLSAALRNR 205
T + +LS A RNR
Sbjct 1193 ATQNPPGLYGGRKVLSRAFRNR 1214
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 38/65 (58%), Gaps = 1/65 (1%)
Query 123 QFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHP 182
+F W D L+AA++ G W++L + +L S SVL+ LNA + G + + E G +V+ H
Sbjct 1802 EFAWRDGPLLAALKAGHWVVLDELNLASQSVLEGLNACFDHRGEIYVPELGMSFQVQ-HE 1860
Query 183 NFRIL 187
+I
Sbjct 1861 KTKIF 1865
Score = 33.5 bits (75), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 28/67 (41%), Gaps = 3/67 (4%)
Query 123 QFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHP 182
+F W L A G W+LL D V+ L LLE+G L+ G +K P
Sbjct 380 EFVWQPGTLTQAATMGHWILLEDIDYAPLDVVSVLIPLLENGELLIPGRGDC---LKVAP 436
Query 183 NFRILLT 189
F+ T
Sbjct 437 GFQFFAT 443
> mmu:100019 Mdn1, 4833432B22Rik, A130070M06, AA958993, D4Abb1e,
Gm135; midasin homolog (yeast); K14572 midasin
Length=5582
Score = 83.2 bits (204), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 47/117 (40%), Positives = 66/117 (56%), Gaps = 8/117 (6%)
Query 93 AGLAHVLESVSSSSAAAAAAAAACSQEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSAS 152
A A ++E S +A+ S FEWVDS LV A++ G WLL+ + + C+ S
Sbjct 2189 ADFAKLVEDFRSFGVKLLQSASGRSH---GSFEWVDSMLVQALKSGDWLLMDNVNFCNPS 2245
Query 153 VLDRLNALLEDGGSLLLTE----GGAPREVKRHPNFRILLTAEAANTHLLSAALRNR 205
VLDRLNALLE GG L + E G+ V +PNFR+ L+ + + +S A+RNR
Sbjct 2246 VLDRLNALLEPGGVLTINERGMVDGSTCTVTPNPNFRLFLSMDPIHGE-ISRAMRNR 2301
Score = 53.1 bits (126), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 32/83 (38%), Positives = 46/83 (55%), Gaps = 8/83 (9%)
Query 128 DSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKR-HPNFRI 186
+ L+ A+RKG W++L + +L VL+ LN LL+D LL+TE +EV R HP F +
Sbjct 1131 EGVLIDAMRKGYWIVLDELNLAPTDVLEALNRLLDDNRELLITET---QEVVRAHPRFML 1187
Query 187 LLTAEAANTH----LLSAALRNR 205
T + +LS A RNR
Sbjct 1188 FATQNPPGLYGGRKVLSRAFRNR 1210
Score = 52.8 bits (125), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 30/86 (34%), Positives = 48/86 (55%), Gaps = 4/86 (4%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLE-DGGSLLLTEGGAPREVKRHP 182
F +V+ L A++KG+W+LL + +L + L+ L+ LLE GSL+L + G + RHP
Sbjct 812 FAFVEGTLAQAIKKGEWILLDEINLAAPETLECLSGLLEGSSGSLVLLDRGDTEPLVRHP 871
Query 183 NFRI---LLTAEAANTHLLSAALRNR 205
+FR+ + A L +RNR
Sbjct 872 DFRLFACMNPATDVGKRNLPPGIRNR 897
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 38/94 (40%), Positives = 50/94 (53%), Gaps = 14/94 (14%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAP----REVK 179
FEW D LV A+++ + LL + L SVL+RLN++LE L+L E G+P EV+
Sbjct 1443 FEWHDGPLVLAMKEDSFFLLDEISLADDSVLERLNSVLEVEKCLVLAEKGSPESKDNEVE 1502
Query 180 -----RHPNFRILLTAEAA---NTHLLSAALRNR 205
+H FRIL T LS ALRNR
Sbjct 1503 LLTAGKH--FRILATMNPGGDFGKKELSPALRNR 1534
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 39/150 (26%), Positives = 68/150 (45%), Gaps = 20/150 (13%)
Query 57 LSGDNSPAAATTADFAAVQRRRQQRAWRL----LFQGCTLAGLAHVLESVSSSSAAAAAA 112
L G N+ A TA A+ +R RA +L L +G G ++ +++ +S
Sbjct 1712 LDGHNTGDYALTAGTTAMNAQRLLRAAKLNKPILLEGSPGVGKTSLVAALAKASGNTLVR 1771
Query 113 AAACSQEEMS---------------QFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRL 157
Q +++ +F W D L+AA++ G W++L + +L S S+L+ L
Sbjct 1772 INLSEQTDITDLFGADLPVEGGRGGEFAWCDGPLLAALKAGHWVVLDELNLASQSILEGL 1831
Query 158 NALLEDGGSLLLTEGGAPREVKRHPNFRIL 187
NA + G + + E G +V+ H RI
Sbjct 1832 NACFDHRGEIYVPELGMSFQVQ-HEKTRIF 1860
Score = 35.0 bits (79), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 29/67 (43%), Gaps = 3/67 (4%)
Query 123 QFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHP 182
+F W L A KG W+LL D V+ L LLE G L+ G +K P
Sbjct 380 EFVWQPGTLTQAATKGYWILLEDIDYAPLDVVSVLIPLLEHGELLIPGHGDC---LKVAP 436
Query 183 NFRILLT 189
F++ T
Sbjct 437 TFQLFAT 443
> cel:F55F10.1 hypothetical protein; K14572 midasin
Length=4368
Score = 80.9 bits (198), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 40/88 (45%), Positives = 56/88 (63%), Gaps = 2/88 (2%)
Query 123 QFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPRE-VKRH 181
+FEW DS V A G WLL+ D +LCSA+VLDRLN+ LE GG L++ E E ++ H
Sbjct 1776 RFEWTDSVFVDAYLHGDWLLIEDVNLCSAAVLDRLNSCLESGGKLVVAERQNSYEPLEPH 1835
Query 182 PNFRILLTAEAANTHLLSAALRNRCCLV 209
P+FR+ L+ + A +S A+RNR +
Sbjct 1836 PDFRVFLSMD-ARVGEISRAMRNRSVEI 1862
Score = 60.1 bits (144), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 35/90 (38%), Positives = 51/90 (56%), Gaps = 4/90 (4%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPRE-VKRHP 182
FEW D +++A+R G LL+ + L SVL+RLN L E+ +LLL++ G E V+ P
Sbjct 1064 FEWSDGVVISAMRDGVPLLVDEISLAEDSVLERLNPLFEEDRALLLSDAGTDTEVVESKP 1123
Query 183 NFRILLTAEAANTH---LLSAALRNRCCLV 209
F+++ T + LS ALRNR V
Sbjct 1124 GFQMIATMNPGGDYGKKELSKALRNRFTEV 1153
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 34/64 (53%), Gaps = 0/64 (0%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPN 183
F W D ++ A++KG+W+LL + +L S SVL+ LNA + L + E E+ N
Sbjct 1407 FRWEDGPVLKAIKKGEWILLDEMNLASQSVLEGLNACFDHRRVLYIAELNREFEIPATSN 1466
Query 184 FRIL 187
R
Sbjct 1467 CRFF 1470
> ath:AT1G67120 ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding / transcription factor binding; K14572
midasin
Length=5336
Score = 78.2 bits (191), Expect = 2e-14, Method: Composition-based stats.
Identities = 37/92 (40%), Positives = 61/92 (66%), Gaps = 5/92 (5%)
Query 118 QEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTE----GG 173
+++ ++FEWV L+ A+ KG+W++L++A+LC+ +VLDR+N+L+E GS+ + E G
Sbjct 2214 KKQSTKFEWVTGMLIKAIEKGEWVVLKNANLCNPTVLDRINSLVEPCGSITINECGIVNG 2273
Query 174 APREVKRHPNFRILLTAEAANTHLLSAALRNR 205
P V HPNFR+ L+ +S A+RNR
Sbjct 2274 EPVTVVPHPNFRLFLSVNPKFGE-VSRAMRNR 2304
Score = 54.3 bits (129), Expect = 3e-07, Method: Composition-based stats.
Identities = 36/93 (38%), Positives = 49/93 (52%), Gaps = 5/93 (5%)
Query 118 QEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAP-- 175
Q+ + F W D LV A+R G +L+ + L SVL+R+N++LE L L E G P
Sbjct 1460 QKWRAIFVWQDGPLVEAMRAGNIVLVDEISLADDSVLERMNSVLETDRKLSLAEKGGPVL 1519
Query 176 REVKRHPNFRILLTAEAANTH---LLSAALRNR 205
EV H +F +L T + LS ALRNR
Sbjct 1520 EEVVAHEDFFVLATMNPGGDYGKKELSPALRNR 1552
Score = 51.6 bits (122), Expect = 2e-06, Method: Composition-based stats.
Identities = 30/82 (36%), Positives = 43/82 (52%), Gaps = 6/82 (7%)
Query 128 DSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRIL 187
+ ALV AVR G W++L + +L + VL+ LN LL+D L + E + HPNF +
Sbjct 1130 EGALVKAVRGGHWIVLDELNLAPSDVLEALNRLLDDNRELFVPE--LSETISAHPNFMLF 1187
Query 188 LTAEAANTH----LLSAALRNR 205
T + +LS A RNR
Sbjct 1188 ATQNPPTLYGGRKILSRAFRNR 1209
Score = 50.1 bits (118), Expect = 6e-06, Method: Composition-based stats.
Identities = 27/65 (41%), Positives = 38/65 (58%), Gaps = 1/65 (1%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLED-GGSLLLTEGGAPREVKRHP 182
F +V+ A V A+R+G W+LL + +L +L RL +LE GSL L E G + RH
Sbjct 825 FTFVEGAFVTALREGHWVLLDEVNLAPPEILGRLIGVLEGVRGSLCLAERGDVMGIPRHL 884
Query 183 NFRIL 187
NFR+
Sbjct 885 NFRLF 889
Score = 47.8 bits (112), Expect = 3e-05, Method: Composition-based stats.
Identities = 26/71 (36%), Positives = 41/71 (57%), Gaps = 3/71 (4%)
Query 117 SQEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPR 176
S E+M +F W D L+ A+++G W+LL + +L SVL+ LNA+L+ + + E G
Sbjct 1796 SDEDM-KFAWSDGILLQALKEGSWVLLDELNLAPQSVLEGLNAILDHRAQVFIPELGCTF 1854
Query 177 EVKRHPNFRIL 187
E P FR+
Sbjct 1855 ECP--PTFRVF 1863
Score = 34.7 bits (78), Expect = 0.27, Method: Composition-based stats.
Identities = 21/83 (25%), Positives = 38/83 (45%), Gaps = 0/83 (0%)
Query 119 EEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREV 178
++ +F W +L A+ G W++L D + V L++LL S L ++G R
Sbjct 403 DQPGEFRWQPGSLTQAIMNGFWVVLEDIDKAPSDVPLVLSSLLGGSCSFLTSQGEEIRIA 462
Query 179 KRHPNFRILLTAEAANTHLLSAA 201
+ F + T E + +H+ A
Sbjct 463 ETFQLFSTISTPECSVSHIRDAG 485
> pfa:PF14_0326 dynein-related AAA-type ATPase; K14572 midasin
Length=8105
Score = 77.4 bits (189), Expect = 3e-14, Method: Composition-based stats.
Identities = 32/79 (40%), Positives = 53/79 (67%), Gaps = 0/79 (0%)
Query 128 DSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRIL 187
+S + A++KG W++++ H + S+LDRLN+L E+ G +LL E G ++++ H NF+I
Sbjct 3121 ESNFIKAIKKGYWIMIKHLHYSTPSLLDRLNSLFEENGYILLHEFGKKKKIRPHKNFQIF 3180
Query 188 LTAEAANTHLLSAALRNRC 206
LT + + +S ALRNRC
Sbjct 3181 LTLSSQEHYKISKALRNRC 3199
Score = 53.9 bits (128), Expect = 4e-07, Method: Composition-based stats.
Identities = 29/76 (38%), Positives = 44/76 (57%), Gaps = 2/76 (2%)
Query 116 CSQEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAP 175
C S F W D LV++++KG L+ + L +SV++RLN++LE +LLLTE G
Sbjct 2094 CINNLKSLFTWYDGILVSSLKKGHIFLMDEISLVESSVIERLNSVLEYERTLLLTEKGGK 2153
Query 176 --REVKRHPNFRILLT 189
+ +K H NF + T
Sbjct 2154 HVKYIKAHDNFYFIGT 2169
Score = 43.5 bits (101), Expect = 5e-04, Method: Composition-based stats.
Identities = 27/84 (32%), Positives = 42/84 (50%), Gaps = 8/84 (9%)
Query 128 DSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRIL 187
+ V AV+ G W++L + +L + VL+ LN +L+D L + E VK H +F +
Sbjct 1669 EGVFVKAVKYGHWVVLDELNLAPSEVLESLNRILDDNKELYIPE--LKSYVKAHKDFMLF 1726
Query 188 LTAEAANTHL------LSAALRNR 205
T AN + LS A R+R
Sbjct 1727 ATQNPANNNSYLGRKELSKAFRSR 1750
Score = 43.5 bits (101), Expect = 6e-04, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 36/65 (55%), Gaps = 2/65 (3%)
Query 123 QFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHP 182
Q+ W D L+ ++KG W+L+ + +L + L+ LN++L+ + + E VK H
Sbjct 2580 QYAWEDGKLIDCMKKGYWILIDEINLANQQTLEGLNSILDHRKEIFIPEKN--EVVKCHK 2637
Query 183 NFRIL 187
NFR+
Sbjct 2638 NFRLF 2642
Score = 41.6 bits (96), Expect = 0.002, Method: Composition-based stats.
Identities = 20/70 (28%), Positives = 35/70 (50%), Gaps = 2/70 (2%)
Query 120 EMSQ--FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPRE 177
EM++ F++ D L+ ++ G W+LL + +L +L RL LL+ +
Sbjct 1166 EMNELIFKFHDGILIDCIKNGHWILLDEINLAQTEILQRLQGLLDSSNKYFDIIEKGNEQ 1225
Query 178 VKRHPNFRIL 187
+K H NFR+
Sbjct 1226 IKVHKNFRLF 1235
Score = 29.6 bits (65), Expect = 8.6, Method: Composition-based stats.
Identities = 17/71 (23%), Positives = 35/71 (49%), Gaps = 3/71 (4%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPN 183
FE+ L A+ G+W++ + + S S++++L + G + ++E G + H +
Sbjct 614 FEYKYGILSKAMMDGKWIVFENINTISNSIIEKLEE-VNSKGYIYISEKG--EYIYPHKD 670
Query 184 FRILLTAEAAN 194
FR+ T N
Sbjct 671 FRLFATITLCN 681
> tgo:TGME49_042620 hypothetical protein ; K14572 midasin
Length=3661
Score = 62.8 bits (151), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 35/86 (40%), Positives = 47/86 (54%), Gaps = 4/86 (4%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPRE-VKRHP 182
FEW D ++ A+++GQ LL + L +VL+RLN LLED ++L+E G E +K P
Sbjct 2601 FEWEDGPIITAMKQGQMFLLDEISLAQDAVLERLNCLLEDSREIVLSEKGGDVEIIKAAP 2660
Query 183 NFRILLTAEAA---NTHLLSAALRNR 205
FR T LS ALRNR
Sbjct 2661 GFRFFATMNPGGDFGKRELSPALRNR 2686
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 37/115 (32%), Positives = 53/115 (46%), Gaps = 16/115 (13%)
Query 89 GCTLAGLAHVLESVSSSSAAAAAAAAACSQEEMS---------------QFEWVDSALVA 133
G T LA L SV + A+A A S ++ +F + LV
Sbjct 1471 GPTPPSLASYLSSVGLKTDASALAGTPTSSHAVTSNTGGGKANGMRSPIRFSLREGLLVD 1530
Query 134 AVRKGQWLLLRDAHLCSASVLDRLNALLEDGG-SLLLTEGGAPREVKRHPNFRIL 187
A+R G+WLLL + +L A VL RL +L+D L+ E + V+ HPNFR+
Sbjct 1531 AIRTGKWLLLDEINLAPADVLQRLLGILDDPKEPFLVLEDSSRAPVQPHPNFRLF 1585
Score = 46.6 bits (109), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 20/41 (48%), Positives = 28/41 (68%), Gaps = 0/41 (0%)
Query 122 SQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLE 162
S+F WVD L AVR G W+LL + +L S + L+ LN+LL+
Sbjct 3233 SRFRWVDGILTRAVRSGDWILLDEINLASQATLEGLNSLLD 3273
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/100 (31%), Positives = 48/100 (48%), Gaps = 10/100 (10%)
Query 116 CSQEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAP 175
C ++ FE + LV A R+G W+++ + +L + VL+ LN LL+D L + E
Sbjct 1893 CGEKGDIVFE--EGPLVKAAREGSWVVIDELNLAPSEVLEGLNRLLDDNRELFIPETNT- 1949
Query 176 REVKRHPNFRILLT------AEAANTHLLSAALRNRCCLV 209
VK H +F++ T + LS A RNR V
Sbjct 1950 -TVKAHADFQLFATQNPSCLGQYGGRKNLSRAFRNRFVEV 1988
> cpv:cgd5_3110 MDN1, midasin ; K14572 midasin
Length=2893
Score = 57.0 bits (136), Expect = 5e-08, Method: Composition-based stats.
Identities = 36/111 (32%), Positives = 55/111 (49%), Gaps = 23/111 (20%)
Query 118 QEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLED------------GG 165
+++ + FEW D ++ A++ G +LLL + +LC SV++RLN+LLED
Sbjct 1903 KKDQALFEWQDGPIIKAMKTGSFLLLDEINLCDDSVIERLNSLLEDQFIKTESGLSYKSR 1962
Query 166 SLLLTEGGAPRE--------VKRHPNFRILLTAEAANTH---LLSAALRNR 205
+ LTE G + H +FRI T + + LS ALRNR
Sbjct 1963 VIYLTEKGGASSFNNNEDYIIHSHKDFRIFATMNPSGDYGKRELSPALRNR 2013
Score = 51.6 bits (122), Expect = 2e-06, Method: Composition-based stats.
Identities = 29/88 (32%), Positives = 47/88 (53%), Gaps = 7/88 (7%)
Query 123 QFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHP 182
+ E+V+ LV ++R G WL+L + +L + VL+ LN LL+ + + E G +K H
Sbjct 1435 ELEFVEGPLVNSIRNGYWLVLDELNLAPSEVLESLNRLLDSNREIFIPETG--EVIKAHE 1492
Query 183 NFRILLTAEAANT-----HLLSAALRNR 205
+F + T A + +LS A RNR
Sbjct 1493 DFMLFATQNPAGSIYGGRKVLSQAFRNR 1520
Score = 42.7 bits (99), Expect = 0.001, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 39/64 (60%), Gaps = 2/64 (3%)
Query 126 WVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFR 185
W+D L+ A++ G W++L + +L + +L+ LN++++ ++ + E G + + H NFR
Sbjct 2447 WIDGILLYAMKNGDWVILDELNLATQQILEGLNSVMDHRRNIYIPEIG--QTIICHENFR 2504
Query 186 ILLT 189
I T
Sbjct 2505 IFAT 2508
Score = 38.9 bits (89), Expect = 0.012, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 30/48 (62%), Gaps = 0/48 (0%)
Query 117 SQEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDG 164
++ + FE++D L+ A+++G WL+L + +L S +L RL +L G
Sbjct 979 NERQKHYFEFLDGILLKAIKEGHWLILDEINLASIDILQRLIPILNRG 1026
> tpv:TP03_0591 hypothetical protein; K14572 midasin
Length=1762
Score = 43.1 bits (100), Expect = 6e-04, Method: Composition-based stats.
Identities = 27/110 (24%), Positives = 48/110 (43%), Gaps = 19/110 (17%)
Query 119 EEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAP--- 175
+ F W + L+ ++ KG W L + +L S+L+++N++LE ++L G P
Sbjct 1403 DSFPNFIWKNGKLIESMIKGNWFLFDEINLVQDSILEKINSILEFNSYIILNSGSRPCNN 1462
Query 176 -------------REVKRHPNFRILLTAEAAN---THLLSAALRNRCCLV 209
V + NFR++ T N LS ++ NR L+
Sbjct 1463 AVSGPQPNRVNDMEYVYSNKNFRVIGTMNPGNDFGKKELSPSISNRFTLI 1512
Score = 41.2 bits (95), Expect = 0.003, Method: Composition-based stats.
Identities = 24/80 (30%), Positives = 44/80 (55%), Gaps = 7/80 (8%)
Query 131 LVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRILLTA 190
LV A++ G W++L + +L S++VL+ LN +L+D + ++E +K H NF + T
Sbjct 1074 LVIAMKYGYWVILDELNLASSNVLECLNRILDDNKEIYISETN--EVIKCHENFMLFATQ 1131
Query 191 EAANT-----HLLSAALRNR 205
++ LS + +NR
Sbjct 1132 NPCDSIYGGRKQLSTSFKNR 1151
> cel:F11C1.5 hypothetical protein
Length=1804
Score = 40.8 bits (94), Expect = 0.003, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 39/77 (50%), Gaps = 15/77 (19%)
Query 128 DSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPRE---------- 177
DSALV A R GQ L++ +A V+ L +LL D G+L+L +G R
Sbjct 774 DSALVKAARSGQILVIDEADKAPLHVIAILKSLL-DTGTLVLGDGRTLRPASSFTEADKT 832
Query 178 ----VKRHPNFRILLTA 190
V HPNFRI++ A
Sbjct 833 NDRIVPIHPNFRIIMLA 849
> dre:561317 si:dkey-18l1.1
Length=1896
Score = 38.9 bits (89), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 24/76 (31%), Positives = 39/76 (51%), Gaps = 12/76 (15%)
Query 126 WVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEG-----------GA 174
+ DS LV AV+ G L++ +A +V L AL+E G ++L +G G
Sbjct 820 YEDSPLVKAVKMGHILVIDEADKAPTNVTCILKALVE-SGEMILADGRRIVSDALEAAGR 878
Query 175 PREVKRHPNFRILLTA 190
P + HP+FR+++ A
Sbjct 879 PNAIPMHPDFRMIVLA 894
> cpv:cgd1_750 dynein heavy chain ; K10413 dynein heavy chain
1, cytosolic
Length=5246
Score = 37.4 bits (85), Expect = 0.037, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 36/68 (52%), Gaps = 13/68 (19%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPN 183
E +S++ A+RKG W++L++ HL S + LD L LL++ R+ + +
Sbjct 4545 LEKAESSIKTAMRKGTWVMLKNVHLSSGN-LDYLEQLLQN------------RQNRPNKG 4591
Query 184 FRILLTAE 191
FR+ L E
Sbjct 4592 FRLFLATE 4599
> dre:569992 dynein, axonemal, heavy chain 5-like
Length=4559
Score = 37.0 bits (84), Expect = 0.050, Method: Composition-based stats.
Identities = 25/73 (34%), Positives = 36/73 (49%), Gaps = 18/73 (24%)
Query 119 EEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREV 178
+E F+ +D+A V +GQWL+L++ HL + D L LE R
Sbjct 4003 QEKVAFQLLDTA----VARGQWLMLQNCHLLVKWLRD-LEKTLE-------------RIT 4044
Query 179 KRHPNFRILLTAE 191
K HP+FR+ LT E
Sbjct 4045 KPHPDFRLWLTTE 4057
> tgo:TGME49_073480 axonemal beta dynein heavy chain, putative
(EC:5.99.1.3)
Length=4273
Score = 35.8 bits (81), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 36/61 (59%), Gaps = 12/61 (19%)
Query 130 ALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRILLT 189
A+ AV G W+LL++ HL SAS L +L ++E+ R+V HP+FR+LLT
Sbjct 3684 AIETAVSTGAWILLQNCHL-SASFLPQLERIVEE---------FPLRDV--HPDFRLLLT 3731
Query 190 A 190
+
Sbjct 3732 S 3732
> pfa:PF10_0224 dynein heavy chain, putative; K06025 [EC:3.6.4.2]
Length=5687
Score = 35.4 bits (80), Expect = 0.14, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 32/63 (50%), Gaps = 12/63 (19%)
Query 129 SALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRILL 188
S L + ++G W++L + HL + + N +LE+ TEG HPNFR L
Sbjct 5135 SKLELSHKEGHWIVLENIHL-----MAKFNLILENVIDKYATEGS-------HPNFRCFL 5182
Query 189 TAE 191
T+E
Sbjct 5183 TSE 5185
> dre:566072 dynein, axonemal, heavy polypeptide 11-like
Length=1584
Score = 35.4 bits (80), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 33/68 (48%), Gaps = 14/68 (20%)
Query 130 ALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRILLT 189
AL A + G W++L++ HL S L RL +LE+ S H N+R+ ++
Sbjct 1043 ALETAAKNGHWVMLQNIHLLS-QWLSRLEEMLENTAS------------NAHSNYRVFMS 1089
Query 190 AE-AANTH 196
E A N H
Sbjct 1090 GEPAPNPH 1097
> cel:T21E12.4 dhc-1; Dynein Heavy Chain family member (dhc-1);
K10413 dynein heavy chain 1, cytosolic
Length=4568
Score = 34.7 bits (78), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 27/83 (32%), Positives = 39/83 (46%), Gaps = 22/83 (26%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCS---ASVLDRLNALLEDGGSLLLTEGGAPREVKR 180
F DSAL A + G+W+LL++ HL A + RL+++ K
Sbjct 4025 FNQADSALGTATKSGRWVLLKNVHLAPSWLAQLEKRLHSM------------------KP 4066
Query 181 HPNFRILLTAEAANTHLLSAALR 203
H FR+ LTAE + L S+ LR
Sbjct 4067 HAQFRLFLTAE-IHPKLPSSILR 4088
> mmu:56087 Dnahc10; dynein, axonemal, heavy chain 10
Length=4591
Score = 34.7 bits (78), Expect = 0.26, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 30/61 (49%), Gaps = 14/61 (22%)
Query 131 LVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRILLTA 190
L AV +GQWL+L++ HL L L+D E R K HP+FR+ LT
Sbjct 4043 LETAVARGQWLMLQNCHL--------LVKWLKD------LEKSLERITKPHPDFRLWLTT 4088
Query 191 E 191
+
Sbjct 4089 D 4089
> hsa:196385 DNAH10, FLJ38262, FLJ43486, FLJ43808, KIAA2017; dynein,
axonemal, heavy chain 10
Length=4471
Score = 34.7 bits (78), Expect = 0.26, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 30/61 (49%), Gaps = 14/61 (22%)
Query 131 LVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRILLTA 190
L AV +GQWL+L++ HL L L+D E R K HP+FR+ LT
Sbjct 3923 LETAVARGQWLMLQNCHL--------LVKWLKD------LEKSLERITKPHPDFRLWLTT 3968
Query 191 E 191
+
Sbjct 3969 D 3969
> tgo:TGME49_094550 dynein heavy chain, putative ; K10413 dynein
heavy chain 1, cytosolic
Length=4937
Score = 34.3 bits (77), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 31/69 (44%), Gaps = 22/69 (31%)
Query 128 DSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKR----HPN 183
+ A+ A R+G W+L ++ HL + + G +++ R HPN
Sbjct 4272 EKAIGQASRQGNWVLFKNVHLSTKWL------------------QGLEKQLYRMQGVHPN 4313
Query 184 FRILLTAEA 192
FRI LT EA
Sbjct 4314 FRIFLTMEA 4322
> hsa:1770 DNAH9, DNAH17L, DNEL1, DYH9, Dnahc9, HL-20, HL20, KIAA0357;
dynein, axonemal, heavy chain 9
Length=4486
Score = 34.3 bits (77), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 32/66 (48%), Gaps = 13/66 (19%)
Query 128 DSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRIL 187
++AL A +KG W++L++ HL A L L LE+ HP FR+
Sbjct 3943 EAALDLAAKKGHWVILQNIHLV-AKWLSTLEKKLEEHSE------------NSHPEFRVF 3989
Query 188 LTAEAA 193
++AE A
Sbjct 3990 MSAEPA 3995
> xla:398849 MGC68485; KIAA0564
Length=946
Score = 32.0 bits (71), Expect = 1.6, Method: Composition-based stats.
Identities = 21/74 (28%), Positives = 37/74 (50%), Gaps = 12/74 (16%)
Query 128 DSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEG-----------GAPR 176
DS LV A +KG L++ +A +V L L+E G ++L++G P
Sbjct 738 DSPLVKACKKGHILVIDEADKAPTNVTCILKTLVES-GEMILSDGRRLVADLSKMDARPN 796
Query 177 EVKRHPNFRILLTA 190
+ HP+F++++ A
Sbjct 797 LIAIHPDFKMIVLA 810
> dre:566413 si:dkey-20n10.1
Length=779
Score = 30.8 bits (68), Expect = 3.2, Method: Composition-based stats.
Identities = 21/56 (37%), Positives = 28/56 (50%), Gaps = 2/56 (3%)
Query 112 AAAACSQEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSL 167
AA A EM++F + A V A + +W L H C A LD + LLE G S+
Sbjct 533 AACASGNYEMAKFLLDNGADVNACDQFKWTPLH--HACHAGQLDIIQLLLEAGASV 586
> hsa:92799 SHKBP1, Sb1; SH3KBP1 binding protein 1
Length=707
Score = 30.4 bits (67), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 31/125 (24%), Positives = 41/125 (32%), Gaps = 4/125 (3%)
Query 2 VEAVDAEAAACTTEDAPRAAPEAQQQQQQ-LQQRLAEETAALLKACEKAATTLCSGLSGD 60
+E+ D D P ++ QQ Q++ + L +CS S D
Sbjct 482 LESADGHGGCSAGNDI---GPYGERDDQQVFIQKVVPSASQLFVRLSSTGQRVCSVRSVD 538
Query 61 NSPAAATTADFAAVQRRRQQRAWRLLFQGCTLAGLAHVLESVSSSSAAAAAAAAACSQEE 120
SP A T RR R R L G LA + + A A QE
Sbjct 539 GSPTTAFTVLECEGSRRLGSRPRRYLLTGQANGSLAMWDLTTAMDGLGQAPAGGLTEQEL 598
Query 121 MSQFE 125
M Q E
Sbjct 599 MEQLE 603
Lambda K H
0.317 0.124 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6494887820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40