bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3805_orf5
Length=275
Score E
Sequences producing significant alignments: (Bits) Value
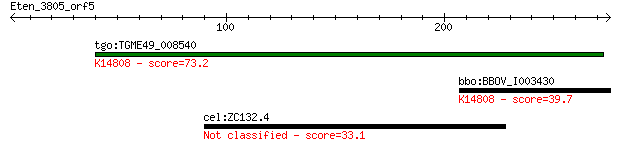
tgo:TGME49_008540 ATP-dependent RNA helicase, putative (EC:5.9... 73.2 9e-13
bbo:BBOV_I003430 19.m02227; DEAD/DEAH box helicase and helicas... 39.7 0.013
cel:ZC132.4 hypothetical protein 33.1 1.1
> tgo:TGME49_008540 ATP-dependent RNA helicase, putative (EC:5.99.1.3);
K14808 ATP-dependent RNA helicase DDX54/DBP10 [EC:3.6.4.13]
Length=983
Score = 73.2 bits (178), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 79/298 (26%), Positives = 135/298 (45%), Gaps = 66/298 (22%)
Query 40 QLLQTMRRFRPKVGVQGSVLSAAAVEAISSRQKE-----------RAAVAAAAAAEERRH 88
++L ++ +RP +G +GSVLSA + + S++ E R + A E+R
Sbjct 551 EVLFRLQNYRPTIGERGSVLSATVMRGMHSKKSEMLRYRNLLQELRESPLENALTEKREE 610
Query 89 RR---AIEDSDADDSDEELLQQQQQGDAGDGSSSSSSGLD------EAQQQQQQ------ 133
+ DAD+SD E ++ G S + + G D EA+ Q+++
Sbjct 611 EEDSPPVSGYDADESDRE-DGLKRSGRVSVTSLNQTPGQDSETSIREAKPQRRRTQESES 669
Query 134 ---------------------QGAGGCNALHALEA--AAAAATAKRRRVSKRRLQKVAKE 170
Q A + L L + T+ ++RVSKRRL++ AKE
Sbjct 670 EEDAQSSSLLNGQASSLGERSQRASASSVLSGLLPRDMTSEGTSAKQRVSKRRLRRTAKE 729
Query 171 MGVDPRAAAATAAAAA---------------AAAAAAAADSSSKSENVFVDVSPEA-KAE 214
MGV +AAA A + + A+ + ++D+ A + +
Sbjct 730 MGVSASSAAAKAQSLSLLQEDREKRDGDRLNGGGDEGEAEGMKRKGGFYLDLQRGACRDK 789
Query 215 GEHVMLQLQHQQHTTYADEEADLRRAKYQEKRKWDPKKRKYVLMRIDSASGLAVKRQK 272
+ +L+L ADE D++R + E+++WDPKKRKY++M++DSA+G AVKR++
Sbjct 790 TQETLLRLDEAAMDLIADENDDMKRQRATEQQRWDPKKRKYIMMKVDSATGRAVKRKR 847
> bbo:BBOV_I003430 19.m02227; DEAD/DEAH box helicase and helicase
conserved C-terminal domain containing protein; K14808 ATP-dependent
RNA helicase DDX54/DBP10 [EC:3.6.4.13]
Length=783
Score = 39.7 bits (91), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 37/70 (52%), Gaps = 1/70 (1%)
Query 207 VSPEAKAEGE-HVMLQLQHQQHTTYADEEADLRRAKYQEKRKWDPKKRKYVLMRIDSASG 265
+ P A E H L+L D E ++++++Q K+ WDPKK+K+V + +D +
Sbjct 565 LVPSAPTNVESHNELKLPQITLNITPDSEELMQKSRFQRKQHWDPKKKKFVQVTVDQMTN 624
Query 266 LAVKRQKKKK 275
+K + +K
Sbjct 625 RVIKNESGQK 634
> cel:ZC132.4 hypothetical protein
Length=831
Score = 33.1 bits (74), Expect = 1.1, Method: Composition-based stats.
Identities = 41/159 (25%), Positives = 66/159 (41%), Gaps = 21/159 (13%)
Query 90 RAIEDSDADDSD--EELLQQ----QQQGDAGDGSSSSSSGLDEAQQQQQQQGAGGCNALH 143
+A++ DA D + EEL QQ Q +GD D S D+A + Q+QG N
Sbjct 487 QAVDQHDAMDIEILEELPQQAPTRQNEGDKQDLQEKDHSRSDQAHKDLQKQGQNKRNHKL 546
Query 144 ALEAAAAAAT---------AKRRRVSKRRLQKVAKEMGVDPRAAAATAAAAAAAAAAAAA 194
A+ A+ A++ ++ RL+K+ KE + A T +A ++ +
Sbjct 547 AVTEASDMMIFHHHQLNEWAQKDPRARSRLEKLQKERQFNVHQALNTLSANKDGTSSKMS 606
Query 195 DSSSKSENVFVDV------SPEAKAEGEHVMLQLQHQQH 227
S K + + + SPE EH + LQ Q
Sbjct 607 LISDKMKEAWTQIQAHMTQSPEPNLRLEHQIQILQKDQQ 645
Lambda K H
0.308 0.117 0.310
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10465222448
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40