bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3821_orf1
Length=211
Score E
Sequences producing significant alignments: (Bits) Value
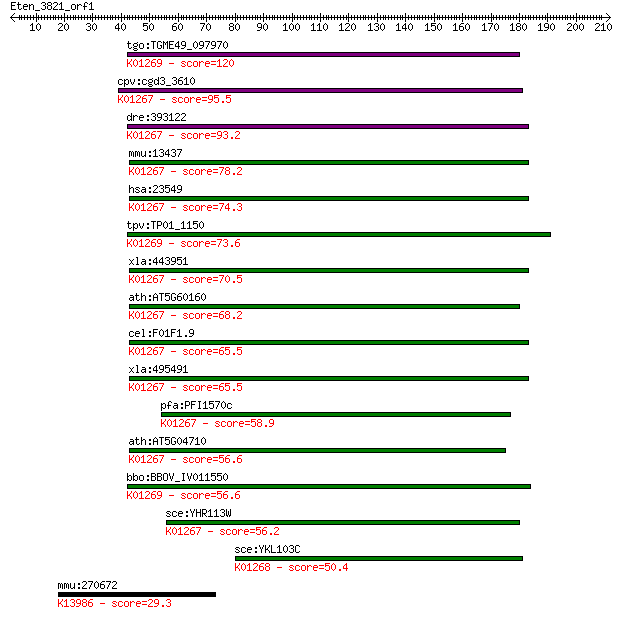
tgo:TGME49_097970 aspartyl aminopeptidase, putative (EC:3.4.11... 120 4e-27
cpv:cgd3_3610 aspartyl aminopeptidase ; K01267 aspartyl aminop... 95.5 1e-19
dre:393122 dnpep, MGC55944, zgc:55944; aspartyl aminopeptidase... 93.2 6e-19
mmu:13437 Dnpep, AA407814; aspartyl aminopeptidase (EC:3.4.11.... 78.2 2e-14
hsa:23549 DNPEP, ASPEP, DAP; aspartyl aminopeptidase (EC:3.4.1... 74.3 3e-13
tpv:TP01_1150 aspartyl aminopeptidase; K01269 aminopeptidase [... 73.6 4e-13
xla:443951 dnpep, MGC80319; aspartyl aminopeptidase (EC:3.4.11... 70.5 4e-12
ath:AT5G60160 aspartyl aminopeptidase, putative; K01267 aspart... 68.2 2e-11
cel:F01F1.9 hypothetical protein; K01267 aspartyl aminopeptida... 65.5 1e-10
xla:495491 hypothetical LOC495491; K01267 aspartyl aminopeptid... 65.5 1e-10
pfa:PFI1570c PfM18AAP; M18 aspartyl aminopeptidase (EC:3.4.11.... 58.9 1e-08
ath:AT5G04710 aspartyl aminopeptidase, putative; K01267 aspart... 56.6 5e-08
bbo:BBOV_IV011550 23.m06108; aspartyl aminopeptidase (EC:3.4.1... 56.6 6e-08
sce:YHR113W Cytoplasmic aspartyl aminopeptidase; cleaves unblo... 56.2 7e-08
sce:YKL103C LAP4, APE1, API, YSC1; Lap4p (EC:3.4.11.22); K0126... 50.4 4e-06
mmu:270672 Map3k15, BC031147, MEKK15; mitogen-activated protei... 29.3 9.9
> tgo:TGME49_097970 aspartyl aminopeptidase, putative (EC:3.4.11.21);
K01269 aminopeptidase [EC:3.4.11.-]
Length=506
Score = 120 bits (301), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 68/158 (43%), Positives = 89/158 (56%), Gaps = 28/158 (17%)
Query 42 QEQHLQPVLCSVI--------------------AEHLNQGNGSSIHEGESEVQRLPPALE 81
+E HLQPVLC+ + A+ +GN S H + + +R L
Sbjct 198 KETHLQPVLCTEVYTQLLAANDSTAREEKRTEEADKETEGNSSCSHLRKRQGERAAAPLL 257
Query 82 RLVMQEIGMPGATLLDWDLCLMDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVETSI 141
L+ QE+ + +++WDLCLMDATPGR G+H EF+ESPRLDNL ST+AAF AL+E
Sbjct 258 SLIAQELRVENEDIVEWDLCLMDATPGRFCGVHEEFVESPRLDNLGSTWAAFSALMECPS 317
Query 142 RQRRGEQDCGETEILMAVAFDHEEVGSESLAGANSSLL 179
EI MAV FDHEE+GSES GA S++L
Sbjct 318 PH--------PEEISMAVGFDHEEIGSESYTGAGSNVL 347
> cpv:cgd3_3610 aspartyl aminopeptidase ; K01267 aspartyl aminopeptidase
[EC:3.4.11.21]
Length=468
Score = 95.5 bits (236), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 53/142 (37%), Positives = 83/142 (58%), Gaps = 18/142 (12%)
Query 39 VFLQEQHLQPVLCSVIAEHLNQGNGSSIHEGESEVQRLPPALERLVMQEIGMPGATLLDW 98
V+ +E+HLQP++ ++ GE ++ LP +L + +EI + + +
Sbjct 168 VYDKEKHLQPIISKT---------DTNDKYGE-QLLSLPRSLLDEICREINVQPQNISSF 217
Query 99 DLCLMDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVETSIRQRRGEQDCGETEILMA 158
DLCLMD+ R GI+ EFI+SPRLDNL F+ F AL++ S + +++L++
Sbjct 218 DLCLMDSVDSRYVGINDEFIDSPRLDNLGGVFSCFTALIDAS--------ESNSSDLLIS 269
Query 159 VAFDHEEVGSESLAGANSSLLE 180
VAFDHEEVGS S +GA+S L+
Sbjct 270 VAFDHEEVGSVSFSGAHSDFLK 291
> dre:393122 dnpep, MGC55944, zgc:55944; aspartyl aminopeptidase
(EC:3.4.11.21); K01267 aspartyl aminopeptidase [EC:3.4.11.21]
Length=469
Score = 93.2 bits (230), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 56/143 (39%), Positives = 83/143 (58%), Gaps = 6/143 (4%)
Query 42 QEQHLQPVLCSVIAEHLNQGNGSSIHEGESE--VQRLPPALERLVMQEIGMPGATLLDWD 99
+E HL P+L + + E L G+ SS + ++ PAL +++ + + LLD++
Sbjct 175 KENHLAPLLATAVQEELETGSASSGDASNATCVAEKHQPALIQMLCGLLSVESNDLLDFE 234
Query 100 LCLMDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVETSIRQRRGEQDCGETEILMAV 159
LCL+D PG L G + EFI SPRLDNL S F A ALV++S + + + M
Sbjct 235 LCLVDTQPGALGGAYEEFIFSPRLDNLHSCFCALTALVDSSTPDSLAK----DPNVRMVT 290
Query 160 AFDHEEVGSESLAGANSSLLELL 182
+D+EEVGSES GA+S+L EL+
Sbjct 291 LYDNEEVGSESAQGAHSNLTELI 313
> mmu:13437 Dnpep, AA407814; aspartyl aminopeptidase (EC:3.4.11.21);
K01267 aspartyl aminopeptidase [EC:3.4.11.21]
Length=475
Score = 78.2 bits (191), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 50/140 (35%), Positives = 76/140 (54%), Gaps = 4/140 (2%)
Query 43 EQHLQPVLCSVIAEHLNQGNGSSIHEGESEVQRLPPALERLVMQEIGMPGATLLDWDLCL 102
E HL P+L + + E L +G G ++ +R L L+ +G+ ++++ +LCL
Sbjct 184 EIHLVPILATAVQEELEKGTPEPGPLGATD-ERHHSVLMSLLCTHLGLSPDSIMEMELCL 242
Query 103 MDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVETSIRQRRGEQDCGETEILMAVAFD 162
D P L G + EFI +PRLDNL S F A +AL+++ +D + M +D
Sbjct 243 ADTQPAVLGGAYEEFIFAPRLDNLHSCFCALQALIDSCASPASLARD---PHVRMVTLYD 299
Query 163 HEEVGSESLAGANSSLLELL 182
+EEVGSES GA S L EL+
Sbjct 300 NEEVGSESAQGAQSLLTELI 319
> hsa:23549 DNPEP, ASPEP, DAP; aspartyl aminopeptidase (EC:3.4.11.21);
K01267 aspartyl aminopeptidase [EC:3.4.11.21]
Length=485
Score = 74.3 bits (181), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 52/146 (35%), Positives = 75/146 (51%), Gaps = 16/146 (10%)
Query 43 EQHLQPVLCSVIAEHLNQGNG-----SSIHEGESEVQRLPPALERLVMQEIGMPGATLLD 97
E HL P+L + I E L +G +++ E V L L+ +G+ +++
Sbjct 194 EMHLVPILATAIQEELEKGTPEPGPLNAVDERHHSV------LMSLLCAHLGLSPKDIVE 247
Query 98 WDLCLMDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVETSIRQRRGEQDCG-ETEIL 156
+LCL D P L G + EFI +PRLDNL S F A +AL+++ G E +
Sbjct 248 MELCLADTQPAVLGGAYDEFIFAPRLDNLHSCFCALQALIDSCA----GPGSLATEPHVR 303
Query 157 MAVAFDHEEVGSESLAGANSSLLELL 182
M +D+EEVGSES GA S L EL+
Sbjct 304 MVTLYDNEEVGSESAQGAQSLLTELV 329
> tpv:TP01_1150 aspartyl aminopeptidase; K01269 aminopeptidase
[EC:3.4.11.-]
Length=457
Score = 73.6 bits (179), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 48/151 (31%), Positives = 81/151 (53%), Gaps = 21/151 (13%)
Query 42 QEQHLQPVLCSVIAEHLNQGNGSSIHEGESEVQRLPPALERLVMQEIGMPGATLLDWDLC 101
++ HL+P++ + + +LN Q P L +L+ E+ L+D++LC
Sbjct 181 KDNHLKPLISTEVVHNLNS------------TQTEP--LLKLISSELDCKVDDLVDFELC 226
Query 102 LMDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVETSIRQRRGEQDCGETEILMAVAF 161
LMD+ P L+G++ EF+ S RLDNL S F + A + + Q GE + +++ V++
Sbjct 227 LMDSNPSCLSGVYEEFVSSGRLDNLGSCFGSISAFTDFVLNQ--GEDN---DAVVVTVSY 281
Query 162 DHEEVGSESLAGANS--SLLELLKASGTLSG 190
++EE+GS GA+S + L L K G L G
Sbjct 282 NYEEIGSSLSYGADSNVTFLWLEKLFGALGG 312
> xla:443951 dnpep, MGC80319; aspartyl aminopeptidase (EC:3.4.11.21);
K01267 aspartyl aminopeptidase [EC:3.4.11.21]
Length=479
Score = 70.5 bits (171), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 50/146 (34%), Positives = 75/146 (51%), Gaps = 9/146 (6%)
Query 43 EQHLQPVLCSVI-----AEHLNQGNGSSIHEGESEV-QRLPPALERLVMQEIGMPGATLL 96
EQ L P+L + + E L+ G + G + + +R P L L+ ++G+ +L
Sbjct 181 EQQLVPILATSVQECLEKETLDSGISCTSASGSNTLTERHHPVLLTLLCDKLGVKPEQIL 240
Query 97 DWDLCLMDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVETSIRQRRGEQDCGETEIL 156
+ +LCL D P L G++ EFI PRLDNL S + A +AL+ + D +
Sbjct 241 EMELCLTDTQPATLGGVYEEFIFGPRLDNLHSCYCALQALLGSCESPSSLASD---PNVR 297
Query 157 MAVAFDHEEVGSESLAGANSSLLELL 182
M +D+EEVGS S GA S L EL+
Sbjct 298 MITLYDNEEVGSGSAQGAESLLTELI 323
> ath:AT5G60160 aspartyl aminopeptidase, putative; K01267 aspartyl
aminopeptidase [EC:3.4.11.21]
Length=477
Score = 68.2 bits (165), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 49/144 (34%), Positives = 68/144 (47%), Gaps = 10/144 (6%)
Query 43 EQHLQPVLCSVIAEHLNQGNGSSIHEGE-------SEVQRLPPALERLVMQEIGMPGATL 95
+ HL PVL + I LN+ S E S + P L ++ +G +
Sbjct 176 QTHLVPVLATAIKAELNKTPAESGEHDEGKKCAETSSKSKHHPLLMEIIANALGCKPEEI 235
Query 96 LDWDLCLMDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVETSIRQRRGEQDCGETEI 155
D++L D P LAG EFI S RLDNL +F + +AL++ + E + G I
Sbjct 236 CDFELQACDTQPSILAGAAKEFIFSGRLDNLCMSFCSLKALIDATSSGSDLEDESG---I 292
Query 156 LMAVAFDHEEVGSESLAGANSSLL 179
M FDHEEVGS S GA S ++
Sbjct 293 RMVALFDHEEVGSNSAQGAGSPVM 316
> cel:F01F1.9 hypothetical protein; K01267 aspartyl aminopeptidase
[EC:3.4.11.21]
Length=470
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 44/143 (30%), Positives = 68/143 (47%), Gaps = 7/143 (4%)
Query 43 EQHLQPVLCSVIAEHLN---QGNGSSIHEGESEVQRLPPALERLVMQEIGMPGATLLDWD 99
E L+P+L + A +N + + + + P L+ +E G ++D D
Sbjct 179 ETELRPILETFAAAGINAPQKPESTGFADPRNITNNHHPQFLGLIAKEAGCQPEDIVDLD 238
Query 100 LCLMDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVETSIRQRRGEQDCGETEILMAV 159
L L D + G+ EFI RLDN T+ A L+E+ GE + +I +A
Sbjct 239 LYLYDTNKAAIVGMEDEFISGARLDNQVGTYTAISGLLES----LTGESFKNDPQIRIAA 294
Query 160 AFDHEEVGSESLAGANSSLLELL 182
FD+EEVGS+S GA+SS E +
Sbjct 295 CFDNEEVGSDSAMGASSSFTEFV 317
> xla:495491 hypothetical LOC495491; K01267 aspartyl aminopeptidase
[EC:3.4.11.21]
Length=479
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 50/146 (34%), Positives = 72/146 (49%), Gaps = 9/146 (6%)
Query 43 EQHLQPVLCSVI-----AEHLNQGNGSSIHEGESEV-QRLPPALERLVMQEIGMPGATLL 96
EQ L P+L S + E L+ G G + + R P L L+ ++G+ +L
Sbjct 181 EQQLLPILASAVQESLEKETLDSGISCPSAPGSNTLADRHHPLLLTLLCDKLGVKPEQIL 240
Query 97 DWDLCLMDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVETSIRQRRGEQDCGETEIL 156
+ +LCL D P L G + EFI PRLDNL S + A +AL+ + + +
Sbjct 241 EMELCLTDTQPATLGGAYEEFIFGPRLDNLHSCYCALQALLGSC---ESSSSLANDPNVR 297
Query 157 MAVAFDHEEVGSESLAGANSSLLELL 182
M +D+EEVGS S GA S L EL+
Sbjct 298 MITLYDNEEVGSGSAQGAESLLTELI 323
> pfa:PFI1570c PfM18AAP; M18 aspartyl aminopeptidase (EC:3.4.11.21);
K01267 aspartyl aminopeptidase [EC:3.4.11.21]
Length=570
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 42/138 (30%), Positives = 60/138 (43%), Gaps = 29/138 (21%)
Query 54 IAEHLNQGNGSSIHEGESEVQRLPPALERLVMQEIGMPGATLLDWDLCLMDATPGRLAGI 113
I EH+N N P L L+ +E+ +LD++LCLMD G+
Sbjct 268 IVEHINTDNSY-------------PLL-YLLSKELNCKEEDILDFELCLMDTQEPCFTGV 313
Query 114 HLEFIESPRLDNLASTFAAFEALVE--TSIRQR-------------RGEQDCGETEILMA 158
+ EFIE R DNL +F FE +E SI+ D + ++
Sbjct 314 YEEFIEGARFDNLLGSFCVFEGFIELVNSIKNHTSNENTNHTNNITNDINDNIHNNLYIS 373
Query 159 VAFDHEEVGSESLAGANS 176
+ +DHEE+GS S GA S
Sbjct 374 IGYDHEEIGSLSEVGARS 391
> ath:AT5G04710 aspartyl aminopeptidase, putative; K01267 aspartyl
aminopeptidase [EC:3.4.11.21]
Length=526
Score = 56.6 bits (135), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 41/132 (31%), Positives = 67/132 (50%), Gaps = 3/132 (2%)
Query 43 EQHLQPVLCSVIAEHLNQGNGSSIHEGESEVQRLPPALERLVMQEIGMPGATLLDWDLCL 102
+ +L+ L ++A ++ + S + S P L +++ ++ ++ +L +
Sbjct 229 KPNLETQLVPLLATKSDESSAESKDKNVSSKDAHHPLLMQILSDDLDCKVEDIVSLELNI 288
Query 103 MDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVETSIRQRRGEQDCGETEILMAVAFD 162
D P L G + EFI S RLDNLAS+F A AL+++ E E +I M FD
Sbjct 289 CDTQPSCLGGANNEFIFSGRLDNLASSFCALRALIDSC---ESSENLSTEHDIRMIALFD 345
Query 163 HEEVGSESLAGA 174
+EEVGS+S GA
Sbjct 346 NEEVGSDSCQGA 357
> bbo:BBOV_IV011550 23.m06108; aspartyl aminopeptidase (EC:3.4.11.21);
K01269 aminopeptidase [EC:3.4.11.-]
Length=429
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 44/151 (29%), Positives = 74/151 (49%), Gaps = 25/151 (16%)
Query 42 QEQHLQPVLCSVIAEHLNQGNGSSIHEGESEVQRLPPALERLVMQEIGMPGATLLDWDLC 101
+E+HL+ V+ + +L NGS P L + +E+G+ ++D DLC
Sbjct 153 KEKHLRGVVATEAVHNL-HSNGSH------------PVLG-FIAKELGVKVEDIVDMDLC 198
Query 102 LMDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVETSIRQRRGE--------QDCGET 153
+ D T L+G++ EF+ S RLDNLAS F+ V+ + G +
Sbjct 199 MFDITKSSLSGLYEEFLSSARLDNLASCFSVLGGFVD--FVKSNGWLLPLITYLKTDYSN 256
Query 154 EILMAVAFDHEEVGSESLAGANSSL-LELLK 183
I + +++EE+GS +GANS + +E +K
Sbjct 257 YITCVIFYNYEEMGSLMASGANSDITIEWIK 287
> sce:YHR113W Cytoplasmic aspartyl aminopeptidase; cleaves unblocked
N-terminal acidic amino acid residues from peptide substrates;
forms a 12 subunit homo-oligomeric complex; M18 metalloprotease
family member; may interact with ribosomes (EC:3.4.11.21);
K01267 aspartyl aminopeptidase [EC:3.4.11.21]
Length=490
Score = 56.2 bits (134), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 43/128 (33%), Positives = 64/128 (50%), Gaps = 15/128 (11%)
Query 56 EHLNQGNGSSIHEGESEVQRLPPALERLVMQEIGMPG-ATLLDWDLCLMDATPGRLAGIH 114
+ +N G +SI ++ VQR L L+ +E+ + + D++L L D L G +
Sbjct 207 KEINNGEFTSI---KTIVQRHHAELLGLIAKELAIDTIEDIEDFELILYDHNASTLGGFN 263
Query 115 LEFIESPRLDNLASTFAAFEALV---ETSIRQRRGEQDCGETEILMAVAFDHEEVGSESL 171
EF+ S RLDNL S F + L +T I + G I + FDHEE+GS S
Sbjct 264 DEFVFSGRLDNLTSCFTSMHGLTLAADTEIDRESG--------IRLMACFDHEEIGSSSA 315
Query 172 AGANSSLL 179
GA+S+ L
Sbjct 316 QGADSNFL 323
> sce:YKL103C LAP4, APE1, API, YSC1; Lap4p (EC:3.4.11.22); K01268
aminopeptidase I [EC:3.4.11.22]
Length=514
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 36/101 (35%), Positives = 51/101 (50%), Gaps = 4/101 (3%)
Query 80 LERLVMQEIGMPGATLLDWDLCLMDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVET 139
L R V + G+ + L+ DL L D G + GI F+ +PRLD+ +FAA AL+
Sbjct 259 LLRYVAKLAGVEVSELIQMDLDLFDVQKGTIGGIGKHFLFAPRLDDRLCSFAAMIALICY 318
Query 140 SIRQRRGEQDCGETEILMAVAFDHEEVGSESLAGANSSLLE 180
+ E D T L +D+EE+GS + GA LLE
Sbjct 319 AKDVNTEESDLFSTVTL----YDNEEIGSLTRQGAKGGLLE 355
> mmu:270672 Map3k15, BC031147, MEKK15; mitogen-activated protein
kinase kinase kinase 15 (EC:2.7.11.25); K13986 mitogen-activated
protein kinase kinase kinase 15 [EC:2.7.11.25]
Length=1331
Score = 29.3 bits (64), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 31/58 (53%), Gaps = 3/58 (5%)
Query 18 VSLHKYSSSCPF-YLQKQSGE-AVFLQEQHLQPVLCSVIAEHLNQGNGSSIH-EGESE 72
+SL K+ C F Y+ S + ++ + CS++ E LN G GS++ EGE++
Sbjct 586 ISLSKFDERCCFLYVHDNSDDFQIYFSTEDQCNRFCSLVKEMLNNGVGSTVELEGEAD 643
Lambda K H
0.319 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6623499460
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40