bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3841_orf1
Length=246
Score E
Sequences producing significant alignments: (Bits) Value
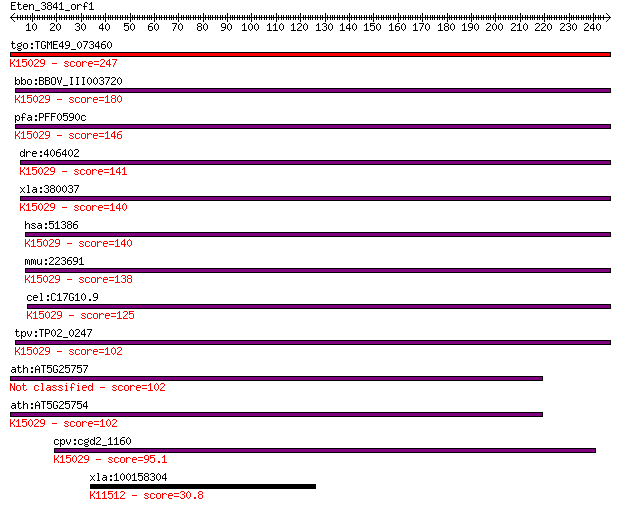
tgo:TGME49_073460 eukaryotic translation initiation factor 3 s... 247 2e-65
bbo:BBOV_III003720 17.m07346; hypothetical protein; K15029 tra... 180 5e-45
pfa:PFF0590c homologue of human HSPC025; K15029 translation in... 146 8e-35
dre:406402 eif3s6ip, wu:fb66c05, zgc:64147; eukaryotic transla... 141 2e-33
xla:380037 eif3l, MGC53371, eif3s6ip; eukaryotic translation i... 140 3e-33
hsa:51386 EIF3L, EIF3EIP, EIF3S11, EIF3S6IP, HSPC025, MSTP005;... 140 6e-33
mmu:223691 Eif3l, 0610011H21Rik, D15N1e, Eif3eip, Eif3ip, Eif3... 138 2e-32
cel:C17G10.9 hypothetical protein; K15029 translation initiati... 125 1e-28
tpv:TP02_0247 hypothetical protein; K15029 translation initiat... 102 9e-22
ath:AT5G25757 hypothetical protein 102 1e-21
ath:AT5G25754 hypothetical protein; K15029 translation initiat... 102 1e-21
cpv:cgd2_1160 HSPC021/HSPC025 family protein ; K15029 translat... 95.1 2e-19
xla:100158304 cenpt, cenp-t; centromere protein T; K11512 cent... 30.8 5.2
> tgo:TGME49_073460 eukaryotic translation initiation factor 3
subunit 6 interacting protein, putative ; K15029 translation
initiation factor 3 subunit L
Length=639
Score = 247 bits (631), Expect = 2e-65, Method: Compositional matrix adjust.
Identities = 122/251 (48%), Positives = 172/251 (68%), Gaps = 6/251 (2%)
Query 1 RHEATWEHRRDSWNNYALLVEFLADTTL----AKDRETITLPLQWVWDMLDEFVYQFQET 56
+ E TWE RR +W NYALL+ FLA+ KD +++ LPLQW+WD+LDEFVYQFQE
Sbjct 151 QQEITWEDRRLAWENYALLINFLAEVECRPDDQKDGKSLRLPLQWLWDILDEFVYQFQEA 210
Query 57 SRWLQRRCKSLA-DSPDRKRLIAELEGDLEVWPAFRVFELLQKVVQQSGIKEQLQQQRAD 115
RW QR + L D +R + E++ D +VW A V E L +VQ+S +++ L+Q + D
Sbjct 211 CRWRQRIARRLPEDEEERALFLKEMDKDCDVWKASPVLEFLHALVQRSEVQDLLRQPKDD 270
Query 116 PMAPKAAETELGFQLGYFALILQLKLHVQVGLFHAGVAAVNDIELSPKAFYWRVPVAHYS 175
K E+E+ FQLGYFA+IL L+LHV +G + GV +V IEL+ + FYW+VP AH +
Sbjct 271 ATKNKLFESEIRFQLGYFAMILLLRLHVLMGDYFMGVKSVESIELASRGFYWKVPAAHVT 330
Query 176 FIFHLGFAYMMLRRYQDAIRVMSQALLALSRQRGQLASSSSYQGASMSRSADRMHVLLLL 235
+HLGFAY M+RRY DAIRVMSQ L+ +SRQR L S+ YQ +M+++ D+M++L++L
Sbjct 331 LFYHLGFAYTMMRRYTDAIRVMSQLLVFVSRQRSYL-SAHFYQQMAMNKAVDKMYILVML 389
Query 236 CSKLSGIKLDD 246
C+ L +KLD+
Sbjct 390 CASLCNVKLDE 400
> bbo:BBOV_III003720 17.m07346; hypothetical protein; K15029 translation
initiation factor 3 subunit L
Length=542
Score = 180 bits (456), Expect = 5e-45, Method: Compositional matrix adjust.
Identities = 100/249 (40%), Positives = 144/249 (57%), Gaps = 17/249 (6%)
Query 3 EATWEHRRDSWNNYALLVEFLADTTLAKDRETITL-----PLQWVWDMLDEFVYQFQETS 57
+ T+E R DSW Y+ L+ + D L K + TL P WVWD+LDEFVYQ QE
Sbjct 107 DVTFEDRLDSWEKYSKLLAYFIDE-LEKGEGSDTLNSLVIPASWVWDILDEFVYQLQECC 165
Query 58 RWLQRRCKSLADSPDRKRLIAELEGDLEVWPAFRVFELLQKVVQQSGIKEQLQQQRADPM 117
RW R +S D K L+++ + +W E+LQ++ + + + D +
Sbjct 166 RWRSRISRS-----DSKALVSDAKIH-SIWKVPTALEMLQRLANNTVFQNMSHPSQRDGL 219
Query 118 APKAAETELGFQLGYFALILQLKLHVQVGLFHAGVAAVNDIELSPKAFYWRVPVAHYSFI 177
L FQLGYFA I ++LHV +G +H V IE+S K YW+VP + S
Sbjct 220 EAAG----LSFQLGYFASISLVRLHVLLGDYHTSVRTATSIEISSKFLYWKVPSYYVSLF 275
Query 178 FHLGFAYMMLRRYQDAIRVMSQALLALSRQRGQLASSSSYQGASMSRSADRMHVLLLLCS 237
+HLGFAYMMLRRY D I+V+SQ L+ L++QR LA S SYQ SM++ A++M+++L+LC
Sbjct 276 YHLGFAYMMLRRYSDCIKVLSQILIFLTKQRTYLA-SQSYQQGSMAKLAEKMYLMLILCH 334
Query 238 KLSGIKLDD 246
+ I+LD+
Sbjct 335 TTTKIRLDE 343
> pfa:PFF0590c homologue of human HSPC025; K15029 translation
initiation factor 3 subunit L
Length=656
Score = 146 bits (368), Expect = 8e-35, Method: Compositional matrix adjust.
Identities = 72/254 (28%), Positives = 141/254 (55%), Gaps = 15/254 (5%)
Query 3 EATWEHRRDSWNNYALLVEFLADTTLAKD------RETITLPLQWVWDMLDEFVYQFQET 56
+ T E R++SW+NY L+ F+ TT+ D + +P W++D L E++YQ+Q
Sbjct 99 DITLEDRKNSWDNYKCLLNFI--TTICTDLSGDSNDNVLVMPNVWIYDFLSEYIYQYQSM 156
Query 57 SRWLQRRCKSLADSPDRKRLIAELEGDLEVWPAFRVFELLQKVVQQSGIKEQLQQQRADP 116
+ R + L D + I + + EV+ + VFE+L ++ + ++++
Sbjct 157 CHY---RLQLLKDLEKNEEGILFVTRNTEVFESSIVFEVLHNLLLKGEFSTLPLEEKSTF 213
Query 117 MAPKAAETEL----GFQLGYFALILQLKLHVQVGLFHAGVAAVNDIELSPKAFYWRVPVA 172
+ +Q YFA + L L+V +G ++ + +++IEL+ KA YW+V +
Sbjct 214 VNNIYNINNEEINSKYQFAYFACCILLNLYVLIGDYYTALKIISNIELNHKALYWKVTLC 273
Query 173 HYSFIFHLGFAYMMLRRYQDAIRVMSQALLALSRQRGQLASSSSYQGASMSRSADRMHVL 232
H S +++ F+YMML+RY D+I+V+SQ L+ LS+Q+ +++ YQ + + D+M+++
Sbjct 274 HLSIFYNIAFSYMMLKRYNDSIKVLSQILIYLSKQKIHVSNQQKYQQNLIHKLIDKMYLI 333
Query 233 LLLCSKLSGIKLDD 246
+++C L+ +LD+
Sbjct 334 IIICHSLTNTRLDE 347
> dre:406402 eif3s6ip, wu:fb66c05, zgc:64147; eukaryotic translation
initiation factor 3, subunit 6 interacting protein; K15029
translation initiation factor 3 subunit L
Length=576
Score = 141 bits (356), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 80/246 (32%), Positives = 138/246 (56%), Gaps = 14/246 (5%)
Query 5 TWEHRRDSWNNYALLVEFLADTTLAKDRETITLPLQWVWDMLDEFVYQFQETSRWLQRRC 64
T + R +S+ NY L ++ + A + LP QW+WD++DEF+YQFQ S++ RC
Sbjct 148 TLDQRFESYYNYCNLFNYILN---ADGPAPLELPNQWLWDIIDEFIYQFQSFSQY---RC 201
Query 65 KSLADSPDRKRLIAELEGDLEVWPAFRVFELLQKVVQQSGIKEQLQ--QQRADP--MAPK 120
K+ S + I L + ++W V +L +V +S I QL+ DP +A +
Sbjct 202 KTAKKSEEE---IEFLRNNPKIWNVHSVLNVLHSLVDKSNINRQLEVYTSGGDPESVAGE 258
Query 121 AAETELGFQLGYFALILQLKLHVQVGLFHAGVAAVNDIELSPKAFYWRVPVAHYSFIFHL 180
L LGYF+L+ L+LH +G ++ + + +IEL+ K+ Y RVP + +++
Sbjct 259 YGRHSLYKMLGYFSLVGLLRLHSLLGDYYQAIKVLENIELNKKSMYSRVPECQITTYYYV 318
Query 181 GFAYMMLRRYQDAIRVMSQALLALSRQRGQLASSSSYQGASMSRSADRMHVLLLLCSKLS 240
GFAY+M+RRYQDAIRV + LL + R R + ++Y+ +++ ++MH LL + +
Sbjct 319 GFAYLMMRRYQDAIRVFANILLYIQRTRN-MFQRTTYKYEMINKQNEQMHGLLAIALTMY 377
Query 241 GIKLDD 246
+++D+
Sbjct 378 PMRIDE 383
> xla:380037 eif3l, MGC53371, eif3s6ip; eukaryotic translation
initiation factor 3, subunit L; K15029 translation initiation
factor 3 subunit L
Length=562
Score = 140 bits (354), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 79/246 (32%), Positives = 138/246 (56%), Gaps = 14/246 (5%)
Query 5 TWEHRRDSWNNYALLVEFLADTTLAKDRETITLPLQWVWDMLDEFVYQFQETSRWLQRRC 64
T E R +S+ NY L ++ + A + LP QW+WD++DEF+YQFQ S++ RC
Sbjct 147 TLEQRFESYYNYCNLFNYILN---ADGPAPLELPNQWLWDIIDEFIYQFQSFSQY---RC 200
Query 65 KSLADSPDRKRLIAELEGDLEVWPAFRVFELLQKVVQQSGIKEQLQ--QQRADP--MAPK 120
K+ S + I L + ++W V +L +V +S I QL+ DP +A +
Sbjct 201 KTAKKSEEE---IEFLRSNPKIWNVHSVLNVLHSLVDKSNINRQLEVYTSGGDPESVAGE 257
Query 121 AAETELGFQLGYFALILQLKLHVQVGLFHAGVAAVNDIELSPKAFYWRVPVAHYSFIFHL 180
L LGYF+L+ L+LH +G ++ + + +IEL+ K+ Y RVP + +++
Sbjct 258 YGRHSLYKMLGYFSLVGLLRLHSLLGDYYQAIKVLENIELNKKSMYSRVPECQVTTYYYV 317
Query 181 GFAYMMLRRYQDAIRVMSQALLALSRQRGQLASSSSYQGASMSRSADRMHVLLLLCSKLS 240
GFAY+M+RRYQD+IRV + LL + R + + ++Y+ +++ ++MH LL + +
Sbjct 318 GFAYLMMRRYQDSIRVFANILLYIQRTKS-MFQRTTYKYEMINKQNEQMHALLSIALTMY 376
Query 241 GIKLDD 246
+++D+
Sbjct 377 PMRIDE 382
> hsa:51386 EIF3L, EIF3EIP, EIF3S11, EIF3S6IP, HSPC025, MSTP005;
eukaryotic translation initiation factor 3, subunit L; K15029
translation initiation factor 3 subunit L
Length=564
Score = 140 bits (352), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 79/244 (32%), Positives = 137/244 (56%), Gaps = 14/244 (5%)
Query 7 EHRRDSWNNYALLVEFLADTTLAKDRETITLPLQWVWDMLDEFVYQFQETSRWLQRRCKS 66
E R +S+ NY L ++ + A + LP QW+WD++DEF+YQFQ S++ RCK+
Sbjct 151 EQRFESYYNYCNLFNYILN---ADGPAPLELPNQWLWDIIDEFIYQFQSFSQY---RCKT 204
Query 67 LADSPDRKRLIAELEGDLEVWPAFRVFELLQKVVQQSGIKEQLQ--QQRADP--MAPKAA 122
S + I L + ++W V +L +V +S I QL+ DP +A +
Sbjct 205 AKKSEEE---IDFLRSNPKIWNVHSVLNVLHSLVDKSNINRQLEVYTSGGDPESVAGEYG 261
Query 123 ETELGFQLGYFALILQLKLHVQVGLFHAGVAAVNDIELSPKAFYWRVPVAHYSFIFHLGF 182
L LGYF+L+ L+LH +G ++ + + +IEL+ K+ Y RVP + +++GF
Sbjct 262 RHSLYKMLGYFSLVGLLRLHSLLGDYYQAIKVLENIELNKKSMYSRVPECQVTTYYYVGF 321
Query 183 AYMMLRRYQDAIRVMSQALLALSRQRGQLASSSSYQGASMSRSADRMHVLLLLCSKLSGI 242
AY+M+RRYQDAIRV + LL + R + + ++Y+ +++ ++MH LL + + +
Sbjct 322 AYLMMRRYQDAIRVFANILLYIQRTKS-MFQRTTYKYEMINKQNEQMHALLAIALTMYPM 380
Query 243 KLDD 246
++D+
Sbjct 381 RIDE 384
> mmu:223691 Eif3l, 0610011H21Rik, D15N1e, Eif3eip, Eif3ip, Eif3s6ip,
HSP-66Y, MGC37328, PAF67; eukaryotic translation initiation
factor 3, subunit L; K15029 translation initiation factor
3 subunit L
Length=564
Score = 138 bits (348), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 79/244 (32%), Positives = 135/244 (55%), Gaps = 14/244 (5%)
Query 7 EHRRDSWNNYALLVEFLADTTLAKDRETITLPLQWVWDMLDEFVYQFQETSRWLQRRCKS 66
E R +S+ NY L ++ + A + LP QW+WD++DEF+YQFQ S++ RCK+
Sbjct 151 EQRFESYYNYCNLFNYILN---ADGPAPLELPNQWLWDIIDEFIYQFQSFSQY---RCKT 204
Query 67 LADSPDRKRLIAELEGDLEVWPAFRVFELLQKVVQQSGIKEQLQ--QQRADP--MAPKAA 122
S L + +VW V +L +V +S I QL+ DP +A +
Sbjct 205 AKKSEGEMDF---LRSNPKVWNVHSVLNVLHSLVDKSNINRQLEVYTSGGDPESVAGEYG 261
Query 123 ETELGFQLGYFALILQLKLHVQVGLFHAGVAAVNDIELSPKAFYWRVPVAHYSFIFHLGF 182
L LGYF+L+ L+LH +G ++ + + +IEL+ K+ Y RVP + +++GF
Sbjct 262 RHSLYKMLGYFSLVGLLRLHSLLGDYYQAIKVLENIELNKKSMYSRVPECQVTTYYYVGF 321
Query 183 AYMMLRRYQDAIRVMSQALLALSRQRGQLASSSSYQGASMSRSADRMHVLLLLCSKLSGI 242
AY+M+RRYQDAIRV + LL + R + + ++Y+ +++ ++MH LL + + +
Sbjct 322 AYLMMRRYQDAIRVFANILLYIQRTKS-MFQRTTYKYEMINKQNEQMHALLAIALTMYPM 380
Query 243 KLDD 246
++D+
Sbjct 381 RIDE 384
> cel:C17G10.9 hypothetical protein; K15029 translation initiation
factor 3 subunit L
Length=535
Score = 125 bits (314), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 72/246 (29%), Positives = 132/246 (53%), Gaps = 13/246 (5%)
Query 8 HRRDSWNNYALLVEFLADTTLAKDRETITLPLQWVWDMLDEFVYQFQETSRWLQRRCKSL 67
HR +S+ NY E ++ +KD ++LP W+WD++DEFVYQFQ + K
Sbjct 115 HRYESFMNYQ---ELFSELLSSKDPIPLSLPNVWLWDIIDEFVYQFQAFCLYKANPGKRN 171
Query 68 ADSPDRKRLIAELEGDLEVWPAFRVFELLQKVVQQSGIKEQL----QQQRADPMAPKAAE 123
AD + + +E + W + V +L ++ +S I EQL +++ D +A + +
Sbjct 172 ADEVED---LINIEENQNAWNIYPVLNILYSLLSKSQIVEQLKALKEKRNPDSVADEFGQ 228
Query 124 TELGFQLGYFALILQLKLHVQVGLFHAGVAAVNDIELSPKAFYWRVPVAHYSFIFHLGFA 183
++L F+LGYFALI L+ HV +G +H + V +++ PK Y VP + + +GF+
Sbjct 229 SDLYFKLGYFALIGLLRTHVLLGDYHQALKTVQYVDIDPKGIYNTVPTCLVTLHYFVGFS 288
Query 184 YMMLRRYQDAIRVMSQALLALSRQR---GQLASSSSYQGASMSRSADRMHVLLLLCSKLS 240
++M+R Y +A ++ LL + R + Q S ++Q + ++ D++ LL +C +
Sbjct 289 HLMMRNYGEATKMFVNCLLYIQRTKSVQNQQPSKKNFQYDVIGKTWDQLFHLLAICLAIQ 348
Query 241 GIKLDD 246
++D+
Sbjct 349 PQRIDE 354
> tpv:TP02_0247 hypothetical protein; K15029 translation initiation
factor 3 subunit L
Length=544
Score = 102 bits (255), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 82/279 (29%), Positives = 126/279 (45%), Gaps = 71/279 (25%)
Query 3 EATWEHRRDSWNNYA-LLVEFLADTTLAKDRETI---TLPLQWVWDMLDEFVYQFQETSR 58
EA++E R DSWN Y LL F+ + + E + T+P WVWDMLDEFVYQ QE+
Sbjct 107 EASYEDRVDSWNKYVRLLTYFIEECNKPEGSEGLSGLTIPSSWVWDMLDEFVYQLQESCH 166
Query 59 WLQRRCKSLADSPDRKRLIAELEGDLEVWPAFRVFELLQKVVQQSGIKEQLQQQRADPMA 118
W R C S D + L+ L +V +QS ++ L +
Sbjct 167 WKIRFCSSTEDQRNCGYLVCSSSS-----------RALTRVGKQSNVQICLSVFKGCAFG 215
Query 119 PKAAE------------------------TELGFQLGY----FAL---ILQLKLHVQVGL 147
+E T+ G + G+ F L LQ H+ G+
Sbjct 216 YNKSELPTRIFRINLLIETSCVTWRLLYFTDNGLKRGFIPQKFVLEDSSLQCHTHLLPGV 275
Query 148 FHAGVAAVNDIELSPKAFYWRVPVAHYSFIFHLGFAYMMLRRYQDAIRVMSQALLALSRQ 207
V+D ++ P +P+ +M RRY DAIR++SQ L+ L++Q
Sbjct 276 ------RVHDAKVLPI-----IPI-------------IMCRRYNDAIRILSQLLIFLTKQ 311
Query 208 RGQLASSSSYQGASMSRSADRMHVLLLLCSKLSGIKLDD 246
R L S SYQ ++MS+ ++M+++++LC + IKLD+
Sbjct 312 RSYLV-SQSYQQSAMSKQTEKMYLIIILCHVTTRIKLDE 349
> ath:AT5G25757 hypothetical protein
Length=514
Score = 102 bits (255), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 70/225 (31%), Positives = 112/225 (49%), Gaps = 19/225 (8%)
Query 1 RHEATWEHRRDSWNNYALLVEFLADTTLAKDRETITLPLQWVWDMLDEFVYQFQETSRWL 60
R T + R DS++NY L + + + + LP QW+WDM+DEFVYQFQ +
Sbjct 98 RLSPTLKQRIDSYDNYCSLFQVVLHGVV-----NMQLPNQWLWDMVDEFVYQFQS---FC 149
Query 61 QRRCKSLADSPDRKRLIAELEGDLEVWPAFRVFELLQKVVQQSGI-------KEQLQQQR 113
Q R K L + + + IA L + W + V LQ +V++S I K+ L+
Sbjct 150 QFRAK-LKNKTEEE--IALLRQHDKAWNVYGVLNFLQALVEKSCIIQILEHDKDGLEFTE 206
Query 114 ADPMAPKAAETELGFQLGYFALILQLKLHVQVGLFHAGVAAVNDIELSPKAFYWRVPVAH 173
D L LGYF+++ L++H +G +H + + I+++ Y V H
Sbjct 207 TDGYDFSGGSNVLKV-LGYFSMVGLLRVHCLLGDYHTALKWLQPIDITQPGVYTSVIGCH 265
Query 174 YSFIFHLGFAYMMLRRYQDAIRVMSQALLALSRQRGQLASSSSYQ 218
+ I+H GFA +MLRRY DA+R ++ LL + + + S Y+
Sbjct 266 IATIYHYGFATLMLRRYVDAVREFNKILLYIFKTKQYHQKSPQYE 310
> ath:AT5G25754 hypothetical protein; K15029 translation initiation
factor 3 subunit L
Length=514
Score = 102 bits (255), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 70/225 (31%), Positives = 112/225 (49%), Gaps = 19/225 (8%)
Query 1 RHEATWEHRRDSWNNYALLVEFLADTTLAKDRETITLPLQWVWDMLDEFVYQFQETSRWL 60
R T + R DS++NY L + + + + LP QW+WDM+DEFVYQFQ +
Sbjct 98 RLSPTLKQRIDSYDNYCSLFQVVLHGVV-----NMQLPNQWLWDMVDEFVYQFQS---FC 149
Query 61 QRRCKSLADSPDRKRLIAELEGDLEVWPAFRVFELLQKVVQQSGI-------KEQLQQQR 113
Q R K L + + + IA L + W + V LQ +V++S I K+ L+
Sbjct 150 QFRAK-LKNKTEEE--IALLRQHDKAWNVYGVLNFLQALVEKSCIIQILEHDKDGLEFTE 206
Query 114 ADPMAPKAAETELGFQLGYFALILQLKLHVQVGLFHAGVAAVNDIELSPKAFYWRVPVAH 173
D L LGYF+++ L++H +G +H + + I+++ Y V H
Sbjct 207 TDGYDFSGGSNVLKV-LGYFSMVGLLRVHCLLGDYHTALKWLQPIDITQPGVYTSVIGCH 265
Query 174 YSFIFHLGFAYMMLRRYQDAIRVMSQALLALSRQRGQLASSSSYQ 218
+ I+H GFA +MLRRY DA+R ++ LL + + + S Y+
Sbjct 266 IATIYHYGFATLMLRRYVDAVREFNKILLYIFKTKQYHQKSPQYE 310
> cpv:cgd2_1160 HSPC021/HSPC025 family protein ; K15029 translation
initiation factor 3 subunit L
Length=673
Score = 95.1 bits (235), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 70/236 (29%), Positives = 121/236 (51%), Gaps = 14/236 (5%)
Query 19 LVEFLADTTLAK--DRET-ITLPLQWVWDMLDEFVYQFQETSRWLQRRCKSLADSPDRKR 75
L E+ L+ DRE I LP + + ++DEF+YQFQ+T R+ R ++ S +
Sbjct 190 LKEYFESNNLSTNFDREDEICLPPEMIHGIIDEFLYQFQDTCRYRTRVAHVISMSEQGQN 249
Query 76 LIAELEGDLEVWPAFRVFELLQKVVQQSGIK------EQLQQQRADPMAPKAAETELGFQ 129
+ D +VW +F++ Q V S ++ QQ + + +Q
Sbjct 250 DGRAAKVDQDVWRCDIIFQIFQSFVDLSEVRLLMSADSTASQQEHLQVLNSVESFPIRYQ 309
Query 130 LGYFALILQLKLHVQVGLFHAGVAAVNDIELSP-KAFYWRVPVAHYSFIFHLGFAYMMLR 188
LGYF+++ L+LH +G ++ + N I+ + W+V A+ S++++LGF+YMML+
Sbjct 310 LGYFSILGLLRLHALLGSYYIAIEVSNGIDHQIFRPLLWKVQTAYVSYLYYLGFSYMMLQ 369
Query 189 RYQDAIRVMSQALLALSRQRGQLASSSS----YQGASMSRSADRMHVLLLLCSKLS 240
RY DAIR+ Q+ ++ + +SS+ YQ ++ + DRM L+L C LS
Sbjct 370 RYIDAIRIFQQSTVSFASNAMNSNNSSAHSSPYQHDAIGKFIDRMSWLMLFCQLLS 425
> xla:100158304 cenpt, cenp-t; centromere protein T; K11512 centromere
protein T
Length=769
Score = 30.8 bits (68), Expect = 5.2, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 44/94 (46%), Gaps = 2/94 (2%)
Query 34 TITLPLQWVWDMLDEFVYQF--QETSRWLQRRCKSLADSPDRKRLIAELEGDLEVWPAFR 91
T++ PL V + E V + +E+S +R+ L D P ++ L+G +E
Sbjct 56 TVSSPLHTVHSKMKERVRRSLRRESSVNPKRQSLPLTDLPVTRKGARGLQGYVEDLDGIT 115
Query 92 VFELLQKVVQQSGIKEQLQQQRADPMAPKAAETE 125
LL+K++Q + Q+ DP+A + E E
Sbjct 116 PRSLLRKIIQNEPEVSLIVSQKYDPVASTSTEPE 149
Lambda K H
0.324 0.135 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8700848608
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40