bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3842_orf1
Length=125
Score E
Sequences producing significant alignments: (Bits) Value
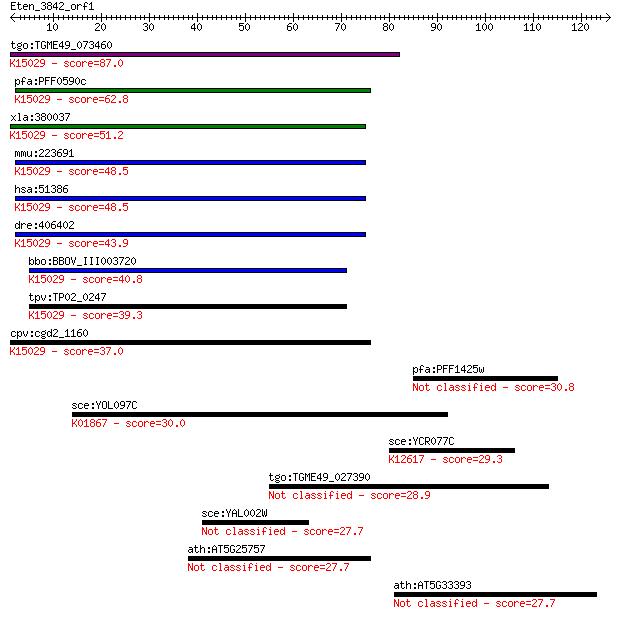
tgo:TGME49_073460 eukaryotic translation initiation factor 3 s... 87.0 1e-17
pfa:PFF0590c homologue of human HSPC025; K15029 translation in... 62.8 3e-10
xla:380037 eif3l, MGC53371, eif3s6ip; eukaryotic translation i... 51.2 9e-07
mmu:223691 Eif3l, 0610011H21Rik, D15N1e, Eif3eip, Eif3ip, Eif3... 48.5 5e-06
hsa:51386 EIF3L, EIF3EIP, EIF3S11, EIF3S6IP, HSPC025, MSTP005;... 48.5 5e-06
dre:406402 eif3s6ip, wu:fb66c05, zgc:64147; eukaryotic transla... 43.9 1e-04
bbo:BBOV_III003720 17.m07346; hypothetical protein; K15029 tra... 40.8 0.001
tpv:TP02_0247 hypothetical protein; K15029 translation initiat... 39.3 0.003
cpv:cgd2_1160 HSPC021/HSPC025 family protein ; K15029 translat... 37.0 0.017
pfa:PFF1425w RNA binding protein, putative 30.8 1.3
sce:YOL097C WRS1, HRE342; Wrs1p (EC:6.1.1.2); K01867 tryptopha... 30.0 2.2
sce:YCR077C PAT1, MRT1; Pat1p; K12617 DNA topoisomerase 2-asso... 29.3 3.6
tgo:TGME49_027390 hypothetical protein 28.9 4.7
sce:YAL002W VPS8, FUN15, VPL8, VPT8; Membrane-associated prote... 27.7 8.5
ath:AT5G25757 hypothetical protein 27.7 8.7
ath:AT5G33393 hypothetical protein 27.7 8.9
> tgo:TGME49_073460 eukaryotic translation initiation factor 3
subunit 6 interacting protein, putative ; K15029 translation
initiation factor 3 subunit L
Length=639
Score = 87.0 bits (214), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 41/81 (50%), Positives = 60/81 (74%), Gaps = 0/81 (0%)
Query 1 PEGEVRSGLMCLKLKSQQKVWRSGPLLSGELTQPHVEALMDFKIDGEMLHVKNQQTQRVY 60
PE VRS ++ +K K +QKVWRSG LLSG+LT + ++F +D +M+HV++QQ+Q+VY
Sbjct 507 PEDAVRSEILSVKYKGRQKVWRSGSLLSGDLTPSSSDNSVEFYMDQDMIHVRSQQSQKVY 566
Query 61 VDYFVKQIERSDALMLAAQHA 81
VD F+KQIERS L + Q++
Sbjct 567 VDQFLKQIERSQHLFNSIQNS 587
> pfa:PFF0590c homologue of human HSPC025; K15029 translation
initiation factor 3 subunit L
Length=656
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 28/74 (37%), Positives = 46/74 (62%), Gaps = 0/74 (0%)
Query 2 EGEVRSGLMCLKLKSQQKVWRSGPLLSGELTQPHVEALMDFKIDGEMLHVKNQQTQRVYV 61
E V S +MC+K S+Q +W+ GPL +G++ DF ID +++++K + Q++++
Sbjct 465 EQNVSSDIMCVKNCSKQLIWKEGPLYTGDIINSIFGNSFDFFIDLDIVNIKTRAHQKIFI 524
Query 62 DYFVKQIERSDALM 75
DYFV QI S LM
Sbjct 525 DYFVHQINMSKNLM 538
> xla:380037 eif3l, MGC53371, eif3s6ip; eukaryotic translation
initiation factor 3, subunit L; K15029 translation initiation
factor 3 subunit L
Length=562
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 41/75 (54%), Gaps = 4/75 (5%)
Query 1 PEGEVRSGLMCLKLKSQQKVWRSG-PLLSGELTQPHVEALMDFKIDGEMLHVKNQQTQRV 59
PE E R L+ K K + VW SG L GE + +DF ID +M+H+ + + R
Sbjct 480 PEQEFRIQLLVFKHKMKNLVWTSGISALEGEFQSA---SEVDFYIDKDMIHIADTKVARR 536
Query 60 YVDYFVKQIERSDAL 74
Y D+F++QI + + L
Sbjct 537 YGDFFIRQIHKFEEL 551
> mmu:223691 Eif3l, 0610011H21Rik, D15N1e, Eif3eip, Eif3ip, Eif3s6ip,
HSP-66Y, MGC37328, PAF67; eukaryotic translation initiation
factor 3, subunit L; K15029 translation initiation factor
3 subunit L
Length=564
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 40/74 (54%), Gaps = 4/74 (5%)
Query 2 EGEVRSGLMCLKLKSQQKVWRSG-PLLSGELTQPHVEALMDFKIDGEMLHVKNQQTQRVY 60
E E R L+ K K + VW SG L GE + +DF ID +M+H+ + + R Y
Sbjct 483 EQEFRIQLLVFKHKMKNLVWTSGISALDGEFQSA---SEVDFYIDKDMIHIADTKVARRY 539
Query 61 VDYFVKQIERSDAL 74
D+F++QI + + L
Sbjct 540 GDFFIRQIHKFEEL 553
> hsa:51386 EIF3L, EIF3EIP, EIF3S11, EIF3S6IP, HSPC025, MSTP005;
eukaryotic translation initiation factor 3, subunit L; K15029
translation initiation factor 3 subunit L
Length=564
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 40/74 (54%), Gaps = 4/74 (5%)
Query 2 EGEVRSGLMCLKLKSQQKVWRSG-PLLSGELTQPHVEALMDFKIDGEMLHVKNQQTQRVY 60
E E R L+ K K + VW SG L GE + +DF ID +M+H+ + + R Y
Sbjct 483 EQEFRIQLLVFKHKMKNLVWTSGISALDGEFQSA---SEVDFYIDKDMIHIADTKVARRY 539
Query 61 VDYFVKQIERSDAL 74
D+F++QI + + L
Sbjct 540 GDFFIRQIHKFEEL 553
> dre:406402 eif3s6ip, wu:fb66c05, zgc:64147; eukaryotic translation
initiation factor 3, subunit 6 interacting protein; K15029
translation initiation factor 3 subunit L
Length=576
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 26/74 (35%), Positives = 40/74 (54%), Gaps = 4/74 (5%)
Query 2 EGEVRSGLMCLKLKSQQKVWRSG-PLLSGELTQPHVEALMDFKIDGEMLHVKNQQTQRVY 60
E E R L+ K K + VW SG L GE + +DF ID +M+H+ + + R Y
Sbjct 482 EQEFRIQLLVFKHKMKNLVWTSGISALDGEFQSA---SEVDFYIDKDMIHIADTKVARRY 538
Query 61 VDYFVKQIERSDAL 74
D+F++QI + + L
Sbjct 539 GDFFIRQIHKFEEL 552
> bbo:BBOV_III003720 17.m07346; hypothetical protein; K15029 translation
initiation factor 3 subunit L
Length=542
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 21/66 (31%), Positives = 42/66 (63%), Gaps = 3/66 (4%)
Query 5 VRSGLMCLKLKSQQKVWRSGPLLSGELTQPHVEALMDFKIDGEMLHVKNQQTQRVYVDYF 64
VRS ++ +K +++Q S G++ E +D ID +L++K++++Q+++VDYF
Sbjct 453 VRSYVLSVKHQTRQFANSSS---GGDVIPGTAEGDIDLYIDQNILYIKSKRSQKLFVDYF 509
Query 65 VKQIER 70
++QI R
Sbjct 510 LQQINR 515
> tpv:TP02_0247 hypothetical protein; K15029 translation initiation
factor 3 subunit L
Length=544
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 38/66 (57%), Gaps = 1/66 (1%)
Query 5 VRSGLMCLKLKSQQKVWRSGPLLSGELTQPHVEALMDFKIDGEMLHVKNQQTQRVYVDYF 64
+RS ++C K +Q V S L L Q + D ID +LH+K+++ Q++Y +YF
Sbjct 453 LRSQILCAKHHVRQFVSNSFSLYHTNLLQTFGDR-DDLYIDQNVLHIKSKKNQKIYAEYF 511
Query 65 VKQIER 70
++QI +
Sbjct 512 LQQINK 517
> cpv:cgd2_1160 HSPC021/HSPC025 family protein ; K15029 translation
initiation factor 3 subunit L
Length=673
Score = 37.0 bits (84), Expect = 0.017, Method: Composition-based stats.
Identities = 26/77 (33%), Positives = 46/77 (59%), Gaps = 4/77 (5%)
Query 1 PEGEVRSGLMCLKLKS-QQKVWRSGPLLSGELTQPHVEALMDFKIDGEMLH-VKNQQTQR 58
P RS ++ +K +S +QK+W+SG L SGEL Q + +D +D + +H ++ +Q +
Sbjct 579 PSSIARSQIIAIKSRSGRQKIWKSGDLHSGELVQ--LNGDVDLLLDLDTIHIIEGKQVDK 636
Query 59 VYVDYFVKQIERSDALM 75
+ D F KQI ++ L+
Sbjct 637 FFGDIFAKQILKARNLL 653
> pfa:PFF1425w RNA binding protein, putative
Length=780
Score = 30.8 bits (68), Expect = 1.3, Method: Composition-based stats.
Identities = 14/30 (46%), Positives = 15/30 (50%), Gaps = 0/30 (0%)
Query 85 PAPQLQQLQPLQPLQQPQQLQQLQQDTPAN 114
P QL QL PL L Q + QL Q P N
Sbjct 422 PLNQLNQLNPLNQLNQLNPMNQLNQLNPMN 451
> sce:YOL097C WRS1, HRE342; Wrs1p (EC:6.1.1.2); K01867 tryptophanyl-tRNA
synthetase [EC:6.1.1.2]
Length=432
Score = 30.0 bits (66), Expect = 2.2, Method: Composition-based stats.
Identities = 24/79 (30%), Positives = 38/79 (48%), Gaps = 9/79 (11%)
Query 14 LKSQQKVWRSGPLLSGELTQPHVEALMDFKIDGEMLHVKNQQTQRVYVD-YFVKQIERSD 72
LK ++SG LLSGE+ + +E L +F VK Q +R VD + +
Sbjct 359 LKECYDKYKSGELLSGEMKKLCIETLQEF--------VKAFQERRAQVDEETLDKFMVPH 410
Query 73 ALMLAAQHAVVAPAPQLQQ 91
L+ + +VAP P+ +Q
Sbjct 411 KLVWGEKERLVAPKPKTKQ 429
> sce:YCR077C PAT1, MRT1; Pat1p; K12617 DNA topoisomerase 2-associated
protein PAT1
Length=796
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 12/26 (46%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 80 HAVVAPAPQLQQLQPLQPLQQPQQLQ 105
+ +APAP QQ+ PLQP+ Q L+
Sbjct 123 QSTMAPAPAPQQMAPLQPILSMQDLE 148
> tgo:TGME49_027390 hypothetical protein
Length=1047
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 29/58 (50%), Gaps = 2/58 (3%)
Query 55 QTQRVYVDYFVKQIERSDALMLAAQHAVVAPAPQLQQLQPLQPLQQPQQLQQLQQDTP 112
T V + YF+ ++ER DA + P P+ ++ QPL L P ++ + DTP
Sbjct 413 DTDTVTLLYFLFEVERGDASRFRFFFQEMVPPPRDERQQPL--LLGPPEIPEFLGDTP 468
> sce:YAL002W VPS8, FUN15, VPL8, VPT8; Membrane-associated protein
that interacts with Vps21p to facilitate soluble vacuolar
protein localization; component of the CORVET complex; required
for localization and trafficking of the CPY sorting receptor;
contains RING finger motif
Length=1274
Score = 27.7 bits (60), Expect = 8.5, Method: Composition-based stats.
Identities = 10/22 (45%), Positives = 17/22 (77%), Gaps = 0/22 (0%)
Query 41 DFKIDGEMLHVKNQQTQRVYVD 62
D+ + E+L VKN++TQ+ Y+D
Sbjct 935 DYNLHQEILEVKNEETQQKYLD 956
> ath:AT5G25757 hypothetical protein
Length=514
Score = 27.7 bits (60), Expect = 8.7, Method: Composition-based stats.
Identities = 11/38 (28%), Positives = 25/38 (65%), Gaps = 0/38 (0%)
Query 38 ALMDFKIDGEMLHVKNQQTQRVYVDYFVKQIERSDALM 75
A +DF I+ +M++V + + Y D+F++QI + + ++
Sbjct 468 ADIDFFINNDMIYVVESKPAKRYGDFFLRQIAKLEGVI 505
> ath:AT5G33393 hypothetical protein
Length=435
Score = 27.7 bits (60), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 20/43 (46%), Positives = 24/43 (55%), Gaps = 1/43 (2%)
Query 81 AVVAPAPQLQQL-QPLQPLQQPQQLQQLQQDTPANARDRRKPQ 122
+V P+ QQ Q L P+Q QQL Q QQ T N RRKP+
Sbjct 388 VIVQPSFASQQAPQRLAPIQVKQQLLQAQQYTWNNDDQRRKPK 430
Lambda K H
0.318 0.131 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2064871684
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40