bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3844_orf2
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
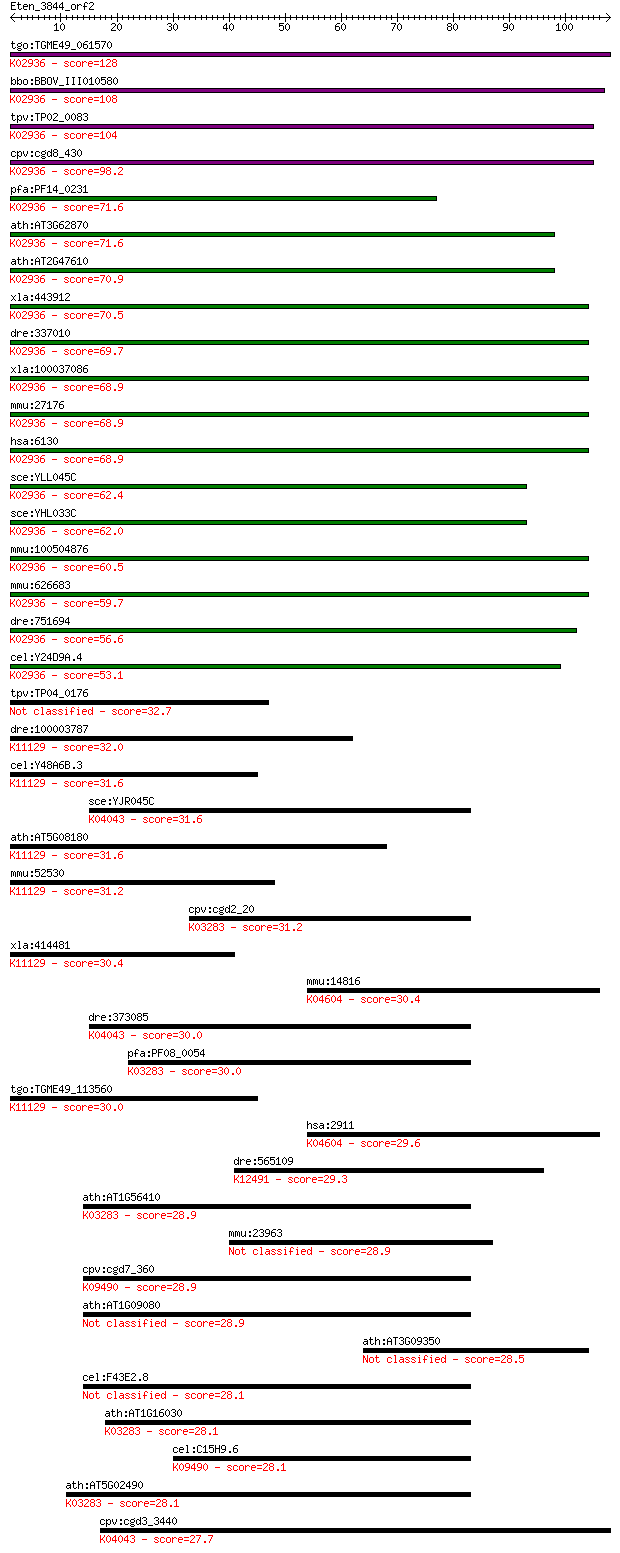
tgo:TGME49_061570 60S ribosomal protein L7a, putative ; K02936... 128 5e-30
bbo:BBOV_III010580 17.m07914; ribosomal protein L7Ae family pr... 108 6e-24
tpv:TP02_0083 60S ribosomal protein L7a; K02936 large subunit ... 104 6e-23
cpv:cgd8_430 60S ribosomal protein L7A ; K02936 large subunit ... 98.2 5e-21
pfa:PF14_0231 60S ribosomal protein L7-3, putative; K02936 lar... 71.6 5e-13
ath:AT3G62870 60S ribosomal protein L7A (RPL7aB); K02936 large... 71.6 5e-13
ath:AT2G47610 60S ribosomal protein L7A (RPL7aA); K02936 large... 70.9 1e-12
xla:443912 rpl7a, MGC80199, surf3, trup; ribosomal protein L7a... 70.5 1e-12
dre:337010 rpl7a, wu:fa95b03, zgc:73183, zgc:86667; ribosomal ... 69.7 2e-12
xla:100037086 hypothetical protein LOC100037086; K02936 large ... 68.9 3e-12
mmu:27176 Rpl7a, MGC103146, Surf3; ribosomal protein L7A; K029... 68.9 3e-12
hsa:6130 RPL7A, SURF3, TRUP; ribosomal protein L7a; K02936 lar... 68.9 3e-12
sce:YLL045C RPL8B, KRB1, SCL41; Rpl8bp; K02936 large subunit r... 62.4 4e-10
sce:YHL033C RPL8A, MAK7; Rpl8ap; K02936 large subunit ribosoma... 62.0 4e-10
mmu:100504876 60S ribosomal protein L7a-like; K02936 large sub... 60.5 1e-09
mmu:626683 Gm11362, OTTMUSG00000000657; predicted gene 11362; ... 59.7 2e-09
dre:751694 MGC153325; zgc:153325; K02936 large subunit ribosom... 56.6 2e-08
cel:Y24D9A.4 rpl-7A; Ribosomal Protein, Large subunit family m... 53.1 2e-07
tpv:TP04_0176 40S ribosomal protein L7Ae 32.7 0.30
dre:100003787 nhp2, MGC73227, nola2, wu:fb36b10, wu:fd61a03, z... 32.0 0.45
cel:Y48A6B.3 hypothetical protein; K11129 H/ACA ribonucleoprot... 31.6 0.59
sce:YJR045C SSC1, ENS1; Hsp70 family ATPase, constituent of th... 31.6 0.67
ath:AT5G08180 ribosomal protein L7Ae/L30e/S12e/Gadd45 family p... 31.6 0.68
mmu:52530 Nhp2, 2410130M07Rik, D11Ertd175e, Nola2; NHP2 ribonu... 31.2 0.76
cpv:cgd2_20 heat shock 70 (HSP70) protein ; K03283 heat shock ... 31.2 0.86
xla:414481 nhp2, MGC81502; NHP2 ribonucleoprotein homolog; K11... 30.4 1.5
mmu:14816 Grm1, 4930455H15Rik, Gm10828, Gprc1a, MGC90744, mGlu... 30.4 1.5
dre:373085 hspa9, MGC86654, cb740, crs, hspa9b, wu:fc14d08, wu... 30.0 1.9
pfa:PF08_0054 heat shock 70 kDa protein; K03283 heat shock 70k... 30.0 2.0
tgo:TGME49_113560 60S ribosomal protein L7a, putative ; K11129... 30.0 2.1
hsa:2911 GRM1, GPRC1A, GRM1A, MGLUR1, MGLUR1A, mGlu1; glutamat... 29.6 2.1
dre:565109 zgc:153779; K12491 Arf-GAP, GTPase, ANK repeat and ... 29.3 3.0
ath:AT1G56410 ERD2; ERD2 (EARLY-RESPONSIVE TO DEHYDRATION 2); ... 28.9 3.7
mmu:23963 Odz1, Odz2, TCAP-1, Ten-m1; odd Oz/ten-m homolog 1 (... 28.9 4.0
cpv:cgd7_360 heat shock protein, Hsp70 ; K09490 heat shock 70k... 28.9 4.0
ath:AT1G09080 BIP3; BIP3; ATP binding 28.9 4.2
ath:AT3G09350 armadillo/beta-catenin repeat family protein 28.5 5.2
cel:F43E2.8 hsp-4; Heat Shock Protein family member (hsp-4) 28.1 6.2
ath:AT1G16030 Hsp70b; Hsp70b (heat shock protein 70B); ATP bin... 28.1 6.6
cel:C15H9.6 hsp-3; Heat Shock Protein family member (hsp-3); K... 28.1 6.7
ath:AT5G02490 heat shock cognate 70 kDa protein 2 (HSC70-2) (H... 28.1 7.7
cpv:cgd3_3440 heat shock protein HSP70, mitochondrial ; K04043... 27.7 8.3
> tgo:TGME49_061570 60S ribosomal protein L7a, putative ; K02936
large subunit ribosomal protein L7Ae
Length=276
Score = 128 bits (321), Expect = 5e-30, Method: Compositional matrix adjust.
Identities = 61/107 (57%), Positives = 77/107 (71%), Gaps = 0/107 (0%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS 60
P++ C +P L RKK+VPY I KGK+RLGQ VH+K C +A+D +KE QAEL AQ K
Sbjct 170 PVELVCFIPQLCRKKEVPYCIVKGKSRLGQLVHQKNCPVLAIDNVRKEDQAELEAQCKIY 229
Query 61 PAMFNDNSALRRRGGGALLGINSQPPQEKKQKLFAPEMRKKAGLQIA 107
AMFNDNS +RRR GG + GI SQ Q+KK+KL E++KK GLQ+A
Sbjct 230 RAMFNDNSEVRRRWGGGINGIKSQHAQQKKEKLINIELKKKMGLQLA 276
> bbo:BBOV_III010580 17.m07914; ribosomal protein L7Ae family
protein; K02936 large subunit ribosomal protein L7Ae
Length=269
Score = 108 bits (269), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 53/106 (50%), Positives = 68/106 (64%), Gaps = 0/106 (0%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS 60
P++ LP L RKK+VPY I KGK+RLG+ VH++T + +AVD +KE QAE K
Sbjct 163 PLELVMWLPTLCRKKEVPYCIIKGKSRLGKLVHQRTASVVAVDNIRKEDQAEFEGMIKAF 222
Query 61 PAMFNDNSALRRRGGGALLGINSQPPQEKKQKLFAPEMRKKAGLQI 106
AMFNDN +RRR GG ++GI SQ + K + L A E KK GL I
Sbjct 223 TAMFNDNVEIRRRWGGHIMGIKSQRQEAKVKALIAMENAKKLGLAI 268
> tpv:TP02_0083 60S ribosomal protein L7a; K02936 large subunit
ribosomal protein L7Ae
Length=266
Score = 104 bits (260), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 52/104 (50%), Positives = 67/104 (64%), Gaps = 0/104 (0%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS 60
PI+ LP L RKK+VPY I K K+RLG+ VH+KT + +AVD +KE QAE +
Sbjct 160 PIELVLWLPALCRKKEVPYCIIKSKSRLGKLVHQKTASVVAVDNVRKEDQAEFDNLVRTF 219
Query 61 PAMFNDNSALRRRGGGALLGINSQPPQEKKQKLFAPEMRKKAGL 104
AMFN+NS LR+R GG ++GI SQ K++ L A E KK GL
Sbjct 220 TAMFNENSELRKRWGGHIMGIKSQHMIAKREALIAMENAKKLGL 263
> cpv:cgd8_430 60S ribosomal protein L7A ; K02936 large subunit
ribosomal protein L7Ae
Length=263
Score = 98.2 bits (243), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 46/104 (44%), Positives = 66/104 (63%), Gaps = 0/104 (0%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS 60
PI+ C LP L R+KDV + I GKARLG+ + KKT A +A++ +KE QAE N
Sbjct 158 PIEIVCFLPALCRRKDVAFCIIGGKARLGKLIGKKTAAVVAIEGVRKEDQAEFDLLCNNF 217
Query 61 PAMFNDNSALRRRGGGALLGINSQPPQEKKQKLFAPEMRKKAGL 104
A +NDN LRR+ GG ++G+ +Q ++++K+ A E KK GL
Sbjct 218 RAQYNDNVELRRKWGGGIMGVKAQHVIKRREKILAMEQAKKVGL 261
> pfa:PF14_0231 60S ribosomal protein L7-3, putative; K02936 large
subunit ribosomal protein L7Ae
Length=283
Score = 71.6 bits (174), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 36/76 (47%), Positives = 45/76 (59%), Gaps = 0/76 (0%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS 60
PI+ LP L R K+VPY I K KA LG+ VHKKT + ++ KKE Q +L +K
Sbjct 176 PIELVLFLPALCRLKEVPYCIVKDKATLGKLVHKKTATAVCLESVKKEDQEKLDYFAKVC 235
Query 61 PAMFNDNSALRRRGGG 76
FNDN LRR+ GG
Sbjct 236 KENFNDNVDLRRKWGG 251
> ath:AT3G62870 60S ribosomal protein L7A (RPL7aB); K02936 large
subunit ribosomal protein L7Ae
Length=256
Score = 71.6 bits (174), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 38/98 (38%), Positives = 54/98 (55%), Gaps = 1/98 (1%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS 60
PI+ LP L RK +VPY I KGK+RLG VH+KT A + + K E + E +
Sbjct 153 PIELVVWLPALCRKMEVPYCIVKGKSRLGAVVHQKTAAALCLTTVKNEDKLEFSKILEAI 212
Query 61 PAMFNDN-SALRRRGGGALLGINSQPPQEKKQKLFAPE 97
A FND R++ GG ++G SQ + K+++ A E
Sbjct 213 KANFNDKYEEYRKKWGGGIMGSKSQAKTKAKERVIAKE 250
> ath:AT2G47610 60S ribosomal protein L7A (RPL7aA); K02936 large
subunit ribosomal protein L7Ae
Length=257
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 37/98 (37%), Positives = 54/98 (55%), Gaps = 1/98 (1%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS 60
PI+ LP L RK +VPY I KGK+RLG VH+KT + + + K E + E +
Sbjct 154 PIELVVWLPALCRKMEVPYCIVKGKSRLGAVVHQKTASCLCLTTVKNEDKLEFSKILEAI 213
Query 61 PAMFNDN-SALRRRGGGALLGINSQPPQEKKQKLFAPE 97
A FND R++ GG ++G SQ + K+++ A E
Sbjct 214 KANFNDKYEEYRKKWGGGIMGSKSQAKTKAKERVIAKE 251
> xla:443912 rpl7a, MGC80199, surf3, trup; ribosomal protein L7a;
K02936 large subunit ribosomal protein L7Ae
Length=266
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 47/107 (43%), Positives = 58/107 (54%), Gaps = 7/107 (6%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKKTC---AFIAVDPFKKEAQAELGAQS 57
PI+ LP L RK VPY I KGKARLG+ VH+KTC AF V+P K A A+L
Sbjct 163 PIELVVFLPALCRKMGVPYCIVKGKARLGRLVHRKTCTSVAFAQVNPEDKGALAKLVEAV 222
Query 58 KNSPAMFNDN-SALRRRGGGALLGINSQPPQEKKQKLFAPEMRKKAG 103
K + +ND +RR GG +LG S K +K A E+ K G
Sbjct 223 KTN---YNDRFDEIRRHWGGGILGPKSVARIAKLEKAKAKELATKLG 266
> dre:337010 rpl7a, wu:fa95b03, zgc:73183, zgc:86667; ribosomal
protein L7a; K02936 large subunit ribosomal protein L7Ae
Length=266
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 43/104 (41%), Positives = 53/104 (50%), Gaps = 1/104 (0%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS 60
PI+ LP L RK VPY I KGKARLG+ VH+KTC IA E +A L +
Sbjct 163 PIELVVFLPALCRKMGVPYCIVKGKARLGRLVHRKTCTSIAFTQTNPEDKAALAKLVEAI 222
Query 61 PAMFNDN-SALRRRGGGALLGINSQPPQEKKQKLFAPEMRKKAG 103
+ND +RR GG +LG S K +K A E+ K G
Sbjct 223 KTNYNDRYEEIRRHWGGNVLGPKSTARIAKLEKAKAKELATKLG 266
> xla:100037086 hypothetical protein LOC100037086; K02936 large
subunit ribosomal protein L7Ae
Length=266
Score = 68.9 bits (167), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 41/104 (39%), Positives = 51/104 (49%), Gaps = 1/104 (0%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS 60
PI+ LP L RK VPY I KGKARLG VH+KTC +A E + L +
Sbjct 163 PIELVVFLPALCRKMGVPYCIIKGKARLGHLVHRKTCTTVAFTQVNSEDKGALAKLVEAI 222
Query 61 PAMFNDN-SALRRRGGGALLGINSQPPQEKKQKLFAPEMRKKAG 103
+ND +RR GG +LG S K +K A E+ K G
Sbjct 223 RTNYNDRYDEIRRHWGGNVLGPKSVARIAKLEKAKAKELATKLG 266
> mmu:27176 Rpl7a, MGC103146, Surf3; ribosomal protein L7A; K02936
large subunit ribosomal protein L7Ae
Length=266
Score = 68.9 bits (167), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 41/104 (39%), Positives = 51/104 (49%), Gaps = 1/104 (0%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS 60
PI+ LP L RK VPY I KGKARLG VH+KTC +A E + L +
Sbjct 163 PIELVVFLPALCRKMGVPYCIIKGKARLGHLVHRKTCTTVAFTQVNSEDKGALAKLVEAI 222
Query 61 PAMFNDN-SALRRRGGGALLGINSQPPQEKKQKLFAPEMRKKAG 103
+ND +RR GG +LG S K +K A E+ K G
Sbjct 223 RTNYNDRYDEIRRHWGGNVLGPKSVARIAKLEKAKAKELATKLG 266
> hsa:6130 RPL7A, SURF3, TRUP; ribosomal protein L7a; K02936 large
subunit ribosomal protein L7Ae
Length=266
Score = 68.9 bits (167), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 41/104 (39%), Positives = 52/104 (50%), Gaps = 1/104 (0%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS 60
PI+ LP L RK VPY I KGKARLG+ VH+KTC +A E + L +
Sbjct 163 PIELVVFLPALCRKMGVPYCIIKGKARLGRLVHRKTCTTVAFTQVNSEDKGALAKLVEAI 222
Query 61 PAMFNDN-SALRRRGGGALLGINSQPPQEKKQKLFAPEMRKKAG 103
+ND +RR GG +LG S K +K A E+ K G
Sbjct 223 RTNYNDRYDEIRRHWGGNVLGPKSVARIAKLEKAKAKELATKLG 266
> sce:YLL045C RPL8B, KRB1, SCL41; Rpl8bp; K02936 large subunit
ribosomal protein L7Ae
Length=256
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 35/93 (37%), Positives = 50/93 (53%), Gaps = 1/93 (1%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS 60
PI+ LP L +K VPY+I KGKARLG V++KT A A+ + E +A L
Sbjct 159 PIELVVFLPALCKKMGVPYAIIKGKARLGTLVNQKTSAVAALTEVRAEDEAALAKLVSTI 218
Query 61 PAMFNDN-SALRRRGGGALLGINSQPPQEKKQK 92
A F D +++ GG +LG +Q +K+ K
Sbjct 219 DANFADKYDEVKKHWGGGILGNKAQAKMDKRAK 251
> sce:YHL033C RPL8A, MAK7; Rpl8ap; K02936 large subunit ribosomal
protein L7Ae
Length=256
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 35/93 (37%), Positives = 50/93 (53%), Gaps = 1/93 (1%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS 60
PI+ LP L +K VPY+I KGKARLG V++KT A A+ + E +A L
Sbjct 159 PIELVVFLPALCKKMGVPYAIVKGKARLGTLVNQKTSAVAALTEVRAEDEAALAKLVSTI 218
Query 61 PAMFNDN-SALRRRGGGALLGINSQPPQEKKQK 92
A F D +++ GG +LG +Q +K+ K
Sbjct 219 DANFADKYDEVKKHWGGGILGNKAQAKMDKRAK 251
> mmu:100504876 60S ribosomal protein L7a-like; K02936 large subunit
ribosomal protein L7Ae
Length=278
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 40/105 (38%), Positives = 50/105 (47%), Gaps = 2/105 (1%)
Query 1 PIDFFCG-LPPLSRKKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKN 59
PI+ LP L RK VPY I KGKARLG VH+KTC +A E + L +
Sbjct 174 PIEMLVVFLPALCRKMGVPYCIIKGKARLGCLVHRKTCTTVAFTRVNSEDKGALAKLVEA 233
Query 60 SPAMFNDN-SALRRRGGGALLGINSQPPQEKKQKLFAPEMRKKAG 103
+ND + R GG +LG S K +K A E+ K G
Sbjct 234 IRTNYNDRYDEIHRHWGGNVLGPKSVACLAKLEKAKAKELAPKLG 278
> mmu:626683 Gm11362, OTTMUSG00000000657; predicted gene 11362;
K02936 large subunit ribosomal protein L7Ae
Length=266
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 37/104 (35%), Positives = 48/104 (46%), Gaps = 1/104 (0%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS 60
PI+ LP L +K VPY I KGKARLG VH+KTC +A E + L +
Sbjct 163 PIELVVFLPALCQKMGVPYCIIKGKARLGHLVHRKTCTTVAFTQVNSENKGALAKLVEAI 222
Query 61 PAMFND-NSALRRRGGGALLGINSQPPQEKKQKLFAPEMRKKAG 103
+ND +R G +LG S K +K A E+ G
Sbjct 223 RTNYNDIYDKIRCHWGDNILGPKSVARIAKLEKAKAKELATNLG 266
> dre:751694 MGC153325; zgc:153325; K02936 large subunit ribosomal
protein L7Ae
Length=269
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 34/102 (33%), Positives = 51/102 (50%), Gaps = 1/102 (0%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS 60
P++ LP L RK VPY I K K+RLG+ V +KTC+ +A+ + + L +
Sbjct 165 PVELVIFLPSLCRKMGVPYCIVKDKSRLGRLVRRKTCSAVALTQVEAGDRNALAKLIETV 224
Query 61 PAMFNDN-SALRRRGGGALLGINSQPPQEKKQKLFAPEMRKK 101
+ND +++ GG LG S K +K A EM +K
Sbjct 225 KTNYNDRFEEIKKHWGGGTLGSKSNARIAKIEKAKAKEMAQK 266
> cel:Y24D9A.4 rpl-7A; Ribosomal Protein, Large subunit family
member (rpl-7A); K02936 large subunit ribosomal protein L7Ae
Length=245
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 34/102 (33%), Positives = 55/102 (53%), Gaps = 7/102 (6%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKKTCAFIA---VDPFKKEAQAELGAQS 57
P++ LP L RK +VPY+I KGKA LG V +KT A +A V+P K A +L
Sbjct 144 PLEIVLHLPALCRKYNVPYAIIKGKASLGTVVRRKTTAAVALVDVNPEDKSALNKLVETV 203
Query 58 KNSPAMFND-NSALRRRGGGALLGINSQPPQEKKQKLFAPEM 98
N+ F++ + +R+ GG ++ S + K ++ A ++
Sbjct 204 NNN---FSERHEEIRKHWGGGVMSAKSDAKKLKIERARARDL 242
> tpv:TP04_0176 40S ribosomal protein L7Ae
Length=177
Score = 32.7 bits (73), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 21/48 (43%), Gaps = 2/48 (4%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKK--TCAFIAVDPFK 46
P+D +P L + + Y+ K L H K TCA + V P K
Sbjct 94 PVDVVSHVPVLCEELSISYAYVASKRVLSDVCHSKRPTCAVLVVKPLK 141
> dre:100003787 nhp2, MGC73227, nola2, wu:fb36b10, wu:fd61a03,
zgc:73227; NHP2 ribonucleoprotein homolog (yeast); K11129 H/ACA
ribonucleoprotein complex subunit 2
Length=150
Score = 32.0 bits (71), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 29/65 (44%), Gaps = 5/65 (7%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKK--TCAFIAVDPFK--KEAQAELGAQ 56
PID +C LP + + +PY+ K LG K TC I + P KEA E +
Sbjct 84 PIDVYCHLPIMCEDRSLPYAYVPSKVDLGSSAGSKRPTCV-IMIKPHDEYKEAYDECVEE 142
Query 57 SKNSP 61
+ P
Sbjct 143 VTSLP 147
> cel:Y48A6B.3 hypothetical protein; K11129 H/ACA ribonucleoprotein
complex subunit 2
Length=163
Score = 31.6 bits (70), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 24/45 (53%), Gaps = 1/45 (2%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFV-HKKTCAFIAVDP 44
PID + +P + +K++PY + +LG V H++ I V P
Sbjct 96 PIDVYSHIPGICEEKEIPYVYIPSREQLGLAVGHRRPSILIFVKP 140
> sce:YJR045C SSC1, ENS1; Hsp70 family ATPase, constituent of
the import motor component of the Translocase of the Inner Mitochondrial
membrane (TIM23 complex); involved in protein translocation
and folding; subunit of SceI endonuclease; K04043
molecular chaperone DnaK
Length=654
Score = 31.6 bits (70), Expect = 0.67, Method: Composition-based stats.
Identities = 25/78 (32%), Positives = 36/78 (46%), Gaps = 10/78 (12%)
Query 15 KDVPYSIFK---GKARL---GQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS----PAMF 64
K VPY I K G A + GQ ++ K+ A+A LG KN+ PA F
Sbjct 115 KQVPYKIVKHSNGDAWVEARGQTYSPAQIGGFVLNKMKETAEAYLGKPVKNAVVTVPAYF 174
Query 65 NDNSALRRRGGGALLGIN 82
ND+ + G ++G+N
Sbjct 175 NDSQRQATKDAGQIVGLN 192
> ath:AT5G08180 ribosomal protein L7Ae/L30e/S12e/Gadd45 family
protein; K11129 H/ACA ribonucleoprotein complex subunit 2
Length=156
Score = 31.6 bits (70), Expect = 0.68, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 28/70 (40%), Gaps = 3/70 (4%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKK---TCAFIAVDPFKKEAQAELGAQS 57
PID LP L + VPY K L Q K C + + P K + AE A+
Sbjct 78 PIDVITHLPILCEEAGVPYVYVPSKEDLAQAGATKRPTCCVLVMLKPAKGDLTAEELAKL 137
Query 58 KNSPAMFNDN 67
K +D+
Sbjct 138 KTDYEQVSDD 147
> mmu:52530 Nhp2, 2410130M07Rik, D11Ertd175e, Nola2; NHP2 ribonucleoprotein
homolog (yeast); K11129 H/ACA ribonucleoprotein
complex subunit 2
Length=153
Score = 31.2 bits (69), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 24/49 (48%), Gaps = 3/49 (6%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKK--TCAFIAVDPFKK 47
PI+ +C LP L +++PY K LG K TC I V P ++
Sbjct 87 PIEVYCHLPVLCEDQNLPYVYIPSKTDLGAATGSKRPTC-VIMVKPHEE 134
> cpv:cgd2_20 heat shock 70 (HSP70) protein ; K03283 heat shock
70kDa protein 1/8
Length=682
Score = 31.2 bits (69), Expect = 0.86, Method: Composition-based stats.
Identities = 17/54 (31%), Positives = 29/54 (53%), Gaps = 4/54 (7%)
Query 33 HKKTCAFIAVDPFKKEAQAELGAQSKNS----PAMFNDNSALRRRGGGALLGIN 82
H + + + + K+ ++A LG Q KN+ PA FND+ + GA+ G+N
Sbjct 127 HAEEISAMVLQKMKEISEAYLGRQIKNAVVTVPAYFNDSQRQATKDAGAIAGLN 180
> xla:414481 nhp2, MGC81502; NHP2 ribonucleoprotein homolog; K11129
H/ACA ribonucleoprotein complex subunit 2
Length=149
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 21/42 (50%), Gaps = 2/42 (4%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQFVHKK--TCAFI 40
PI+ +C +P + + +PYS K+ LG K TC +
Sbjct 83 PIEVYCHIPVMCEDRGIPYSYVPSKSDLGAAAGSKRPTCVIL 124
> mmu:14816 Grm1, 4930455H15Rik, Gm10828, Gprc1a, MGC90744, mGluR1,
nmf373, rcw, wobl; glutamate receptor, metabotropic 1;
K04604 metabotropic glutamate receptor 1/5
Length=906
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 28/52 (53%), Gaps = 5/52 (9%)
Query 54 GAQSKNSPAMFNDNSALRRRGGGALLGINSQPPQEKKQKLFAPEMRKKAGLQ 105
GA S+ S A + + + GAL ++ QPP EK + E+R++ G+Q
Sbjct 31 GASSQRSVARMDGDVII-----GALFSVHHQPPAEKVPERKCGEIREQYGIQ 77
> dre:373085 hspa9, MGC86654, cb740, crs, hspa9b, wu:fc14d08,
wu:fc27c10, wu:fc38a06; heat shock protein 9; K04043 molecular
chaperone DnaK
Length=682
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 25/79 (31%), Positives = 38/79 (48%), Gaps = 12/79 (15%)
Query 15 KDVPYSIFK---GKARL----GQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS----PAM 63
K+VPY I + G A L + + AFI + K+ A++ LG KN+ PA
Sbjct 141 KNVPYKIVRASNGDAWLEVHGKMYSPSQAGAFILIK-MKETAESYLGQSVKNAVVTVPAY 199
Query 64 FNDNSALRRRGGGALLGIN 82
FND+ + G + G+N
Sbjct 200 FNDSQRQATKDAGQIAGLN 218
> pfa:PF08_0054 heat shock 70 kDa protein; K03283 heat shock 70kDa
protein 1/8
Length=677
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 18/65 (27%), Positives = 33/65 (50%), Gaps = 7/65 (10%)
Query 22 FKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS----PAMFNDNSALRRRGGGA 77
++G+ +L H + + + + K+ A+A LG KN+ PA FND+ + G
Sbjct 119 YQGEKKL---FHPEEISSMVLQKMKENAEAFLGKSIKNAVITVPAYFNDSQRQATKDAGT 175
Query 78 LLGIN 82
+ G+N
Sbjct 176 IAGLN 180
> tgo:TGME49_113560 60S ribosomal protein L7a, putative ; K11129
H/ACA ribonucleoprotein complex subunit 2
Length=173
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 22/45 (48%), Gaps = 1/45 (2%)
Query 1 PIDFFCGLPPLSRKKDVPYSIFKGKARLGQ-FVHKKTCAFIAVDP 44
PI+ LP L +KDV Y+ K LG F K+ + I + P
Sbjct 95 PIEIIAHLPILCEEKDVVYAYLCSKKTLGHAFRSKRPASVIMITP 139
> hsa:2911 GRM1, GPRC1A, GRM1A, MGLUR1, MGLUR1A, mGlu1; glutamate
receptor, metabotropic 1; K04604 metabotropic glutamate
receptor 1/5
Length=1194
Score = 29.6 bits (65), Expect = 2.1, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 28/52 (53%), Gaps = 5/52 (9%)
Query 54 GAQSKNSPAMFNDNSALRRRGGGALLGINSQPPQEKKQKLFAPEMRKKAGLQ 105
GA S+ S A + + + GAL ++ QPP EK + E+R++ G+Q
Sbjct 31 GASSQRSVARMDGDVII-----GALFSVHHQPPAEKVPERKCGEIREQYGIQ 77
> dre:565109 zgc:153779; K12491 Arf-GAP, GTPase, ANK repeat and
PH domain-containing protein 1/2/3
Length=827
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 25/58 (43%), Gaps = 3/58 (5%)
Query 41 AVDPFKKEAQAELGAQSKNSPAMFNDNSALRR---RGGGALLGINSQPPQEKKQKLFA 95
A P QA GAQS + P+ + RGG + G+ + P ++ LFA
Sbjct 254 ASTPVSGPGQASNGAQSSDYPSSLPSTPVISHKDIRGGASRDGVTQRNPPRRRTSLFA 311
> ath:AT1G56410 ERD2; ERD2 (EARLY-RESPONSIVE TO DEHYDRATION 2);
ATP binding; K03283 heat shock 70kDa protein 1/8
Length=617
Score = 28.9 bits (63), Expect = 3.7, Method: Composition-based stats.
Identities = 19/73 (26%), Positives = 34/73 (46%), Gaps = 5/73 (6%)
Query 14 KKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS----PAMFNDNSA 69
+ D P K QF ++ + + + ++ A+A LG+ KN+ PA FND+
Sbjct 101 QADKPMIFVNYKGEEKQFAAEEISSMVLIK-MREIAEAYLGSSIKNAVVTVPAYFNDSQR 159
Query 70 LRRRGGGALLGIN 82
+ G + G+N
Sbjct 160 QATKDAGVIAGLN 172
> mmu:23963 Odz1, Odz2, TCAP-1, Ten-m1; odd Oz/ten-m homolog 1
(Drosophila)
Length=2731
Score = 28.9 bits (63), Expect = 4.0, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 25/47 (53%), Gaps = 1/47 (2%)
Query 40 IAVDPFKKEAQAELGAQSKNSPAMFNDNSALRRRGGGALLGINSQPP 86
+ + F+ +A + LG S N +FN S + +G G + I+ QPP
Sbjct 1120 VVLQGFEMDA-SNLGGWSLNKHHIFNPQSGIIHKGNGENMFISQQPP 1165
> cpv:cgd7_360 heat shock protein, Hsp70 ; K09490 heat shock 70kDa
protein 5
Length=655
Score = 28.9 bits (63), Expect = 4.0, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 33/73 (45%), Gaps = 5/73 (6%)
Query 14 KKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS----PAMFNDNSA 69
K PY K Q ++ A + V K+ A+A LG + K++ PA FND
Sbjct 124 KDSKPYIQVSVKGEKKQLAPEEVSAMVLVK-MKEIAEAYLGKEVKHAVITVPAYFNDAQR 182
Query 70 LRRRGGGALLGIN 82
+ GA+ G+N
Sbjct 183 QATKDAGAIAGLN 195
> ath:AT1G09080 BIP3; BIP3; ATP binding
Length=675
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 23/73 (31%), Positives = 34/73 (46%), Gaps = 5/73 (6%)
Query 14 KKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS----PAMFNDNSA 69
K PY K K F ++ A I + K+ A+A LG + K++ PA FND
Sbjct 143 KDGKPYIQVKVKGEEKLFSPEEISAMI-LTKMKETAEAFLGKKIKDAVITVPAYFNDAQR 201
Query 70 LRRRGGGALLGIN 82
+ GA+ G+N
Sbjct 202 QATKDAGAIAGLN 214
> ath:AT3G09350 armadillo/beta-catenin repeat family protein
Length=327
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 23/40 (57%), Gaps = 1/40 (2%)
Query 64 FNDNSALRRRGGGALLGINSQPPQEKKQKLFAPEMRKKAG 103
+N+ S+LR +G L G ++ PP + KLF P +R A
Sbjct 272 YNEPSSLREKGLVVLPGEDALPP-DVASKLFEPPLRASAA 310
> cel:F43E2.8 hsp-4; Heat Shock Protein family member (hsp-4)
Length=657
Score = 28.1 bits (61), Expect = 6.2, Method: Composition-based stats.
Identities = 20/73 (27%), Positives = 35/73 (47%), Gaps = 5/73 (6%)
Query 14 KKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS----PAMFNDNSA 69
K + P K + QF ++ A + + K+ A++ LG + KN+ PA FND
Sbjct 125 KSNKPNVEVKVGSETKQFTPEEVSAMV-LTKMKQIAESYLGHEVKNAVVTVPAYFNDAQK 183
Query 70 LRRRGGGALLGIN 82
+ G++ G+N
Sbjct 184 QATKDAGSIAGLN 196
> ath:AT1G16030 Hsp70b; Hsp70b (heat shock protein 70B); ATP binding;
K03283 heat shock 70kDa protein 1/8
Length=646
Score = 28.1 bits (61), Expect = 6.6, Method: Composition-based stats.
Identities = 21/69 (30%), Positives = 33/69 (47%), Gaps = 5/69 (7%)
Query 18 PYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS----PAMFNDNSALRRR 73
P + K QF ++ + + V K+ A+A LG KN+ PA FND+ +
Sbjct 104 PMIVVSYKNEEKQFSPEEISSMVLVK-MKEVAEAFLGRTVKNAVVTVPAYFNDSQRQATK 162
Query 74 GGGALLGIN 82
GA+ G+N
Sbjct 163 DAGAISGLN 171
> cel:C15H9.6 hsp-3; Heat Shock Protein family member (hsp-3);
K09490 heat shock 70kDa protein 5
Length=661
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 29/57 (50%), Gaps = 5/57 (8%)
Query 30 QFVHKKTCAFIAVDPFKKEAQAELGAQSKNS----PAMFNDNSALRRRGGGALLGIN 82
QF ++ A + V K+ A++ LG + KN+ PA FND + G + G+N
Sbjct 144 QFTPEEVSAMVLVK-MKEIAESYLGKEVKNAVVTVPAYFNDAQRQATKDAGTIAGLN 199
> ath:AT5G02490 heat shock cognate 70 kDa protein 2 (HSC70-2)
(HSP70-2); K03283 heat shock 70kDa protein 1/8
Length=653
Score = 28.1 bits (61), Expect = 7.7, Method: Composition-based stats.
Identities = 19/76 (25%), Positives = 36/76 (47%), Gaps = 5/76 (6%)
Query 11 LSRKKDVPYSIFKGKARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS----PAMFND 66
+S + P + + K QF ++ + + + ++ A+A LG KN+ PA FND
Sbjct 98 ISGTAEKPMIVVEYKGEEKQFAAEEISSMVLIK-MREIAEAFLGTTVKNAVVTVPAYFND 156
Query 67 NSALRRRGGGALLGIN 82
+ + G + G+N
Sbjct 157 SQRQATKDAGVIAGLN 172
> cpv:cgd3_3440 heat shock protein HSP70, mitochondrial ; K04043
molecular chaperone DnaK
Length=683
Score = 27.7 bits (60), Expect = 8.3, Method: Composition-based stats.
Identities = 28/103 (27%), Positives = 47/103 (45%), Gaps = 13/103 (12%)
Query 17 VPYSIFKG-------KARLGQFVHKKTCAFIAVDPFKKEAQAELGAQSKNS----PAMFN 65
+PY I + +AR ++ + AFI ++ K+ A+ LG K++ PA FN
Sbjct 138 LPYKIVRADNGDAWVEARGERYSPSQIGAFI-LEKMKETAETYLGRGVKHAVITVPAYFN 196
Query 66 DNSALRRRGGGALLGIN-SQPPQEKKQKLFAPEMRKKAGLQIA 107
D+ + G++ G+N ++ E A M K G IA
Sbjct 197 DSQRQATKDAGSIAGLNVTRIINEPTAAALAYGMEKADGKTIA 239
Lambda K H
0.319 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2007980300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40