bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3849_orf1
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
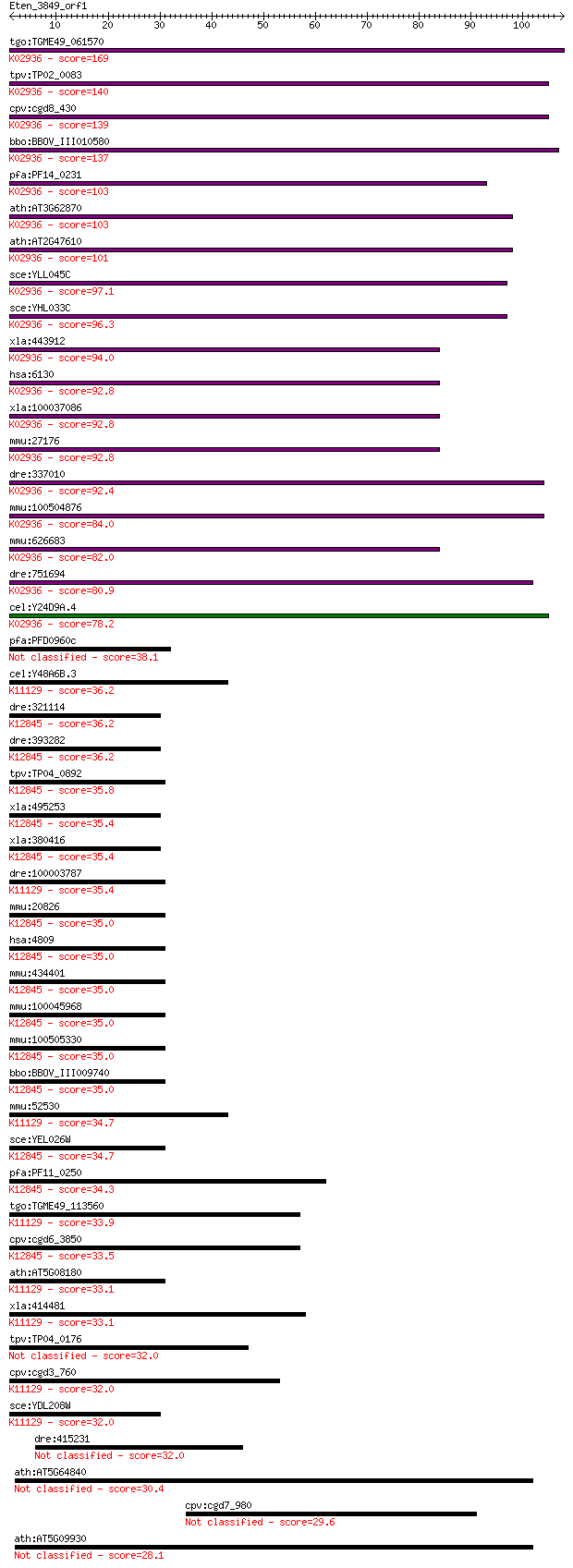
tgo:TGME49_061570 60S ribosomal protein L7a, putative ; K02936... 169 2e-42
tpv:TP02_0083 60S ribosomal protein L7a; K02936 large subunit ... 140 7e-34
cpv:cgd8_430 60S ribosomal protein L7A ; K02936 large subunit ... 139 2e-33
bbo:BBOV_III010580 17.m07914; ribosomal protein L7Ae family pr... 137 6e-33
pfa:PF14_0231 60S ribosomal protein L7-3, putative; K02936 lar... 103 1e-22
ath:AT3G62870 60S ribosomal protein L7A (RPL7aB); K02936 large... 103 2e-22
ath:AT2G47610 60S ribosomal protein L7A (RPL7aA); K02936 large... 101 5e-22
sce:YLL045C RPL8B, KRB1, SCL41; Rpl8bp; K02936 large subunit r... 97.1 1e-20
sce:YHL033C RPL8A, MAK7; Rpl8ap; K02936 large subunit ribosoma... 96.3 2e-20
xla:443912 rpl7a, MGC80199, surf3, trup; ribosomal protein L7a... 94.0 1e-19
hsa:6130 RPL7A, SURF3, TRUP; ribosomal protein L7a; K02936 lar... 92.8 2e-19
xla:100037086 hypothetical protein LOC100037086; K02936 large ... 92.8 3e-19
mmu:27176 Rpl7a, MGC103146, Surf3; ribosomal protein L7A; K029... 92.8 3e-19
dre:337010 rpl7a, wu:fa95b03, zgc:73183, zgc:86667; ribosomal ... 92.4 3e-19
mmu:100504876 60S ribosomal protein L7a-like; K02936 large sub... 84.0 1e-16
mmu:626683 Gm11362, OTTMUSG00000000657; predicted gene 11362; ... 82.0 4e-16
dre:751694 MGC153325; zgc:153325; K02936 large subunit ribosom... 80.9 1e-15
cel:Y24D9A.4 rpl-7A; Ribosomal Protein, Large subunit family m... 78.2 6e-15
pfa:PFD0960c 60S ribosomal protein L7Ae/L30e, putative 38.1 0.007
cel:Y48A6B.3 hypothetical protein; K11129 H/ACA ribonucleoprot... 36.2 0.024
dre:321114 nhp2l1, Ssfa1, cb127, sb:cb127, zgc:56265; NHP2 non... 36.2 0.025
dre:393282 MGC56066, MGC77417, zgc:77417; zgc:56066; K12845 U4... 36.2 0.027
tpv:TP04_0892 hypothetical protein; K12845 U4/U6 small nuclear... 35.8 0.038
xla:495253 nhp2l1-a, fa-1, fa1, hoip, nhp2l1, nhpx, otk27, snr... 35.4 0.038
xla:380416 nhp2l1-b, MGC53430, fa-1, fa1, hoip, nhpx, otk27, s... 35.4 0.038
dre:100003787 nhp2, MGC73227, nola2, wu:fb36b10, wu:fd61a03, z... 35.4 0.045
mmu:20826 Nhp2l1, FA-1, Fta1, Ssfa1; NHP2 non-histone chromoso... 35.0 0.052
hsa:4809 NHP2L1, 15.5K, FA-1, FA1, NHPX, OTK27, SNRNP15-5, SNU... 35.0 0.052
mmu:434401 Gm5616, EG434401; predicted gene 5616; K12845 U4/U6... 35.0 0.053
mmu:100045968 NHP2-like protein 1-like; K12845 U4/U6 small nuc... 35.0 0.055
mmu:100505330 NHP2-like protein 1-like; K12845 U4/U6 small nuc... 35.0 0.062
bbo:BBOV_III009740 17.m07845; ribosomal protein L7A; K12845 U4... 35.0 0.065
mmu:52530 Nhp2, 2410130M07Rik, D11Ertd175e, Nola2; NHP2 ribonu... 34.7 0.066
sce:YEL026W SNU13; RNA binding protein, part of U3 snoRNP invo... 34.7 0.077
pfa:PF11_0250 high mobility group-like protein NHP2, putative;... 34.3 0.089
tgo:TGME49_113560 60S ribosomal protein L7a, putative ; K11129... 33.9 0.11
cpv:cgd6_3850 HOI-POLLOI protein; U4/U6.U5 snRNP component; Sn... 33.5 0.16
ath:AT5G08180 ribosomal protein L7Ae/L30e/S12e/Gadd45 family p... 33.1 0.19
xla:414481 nhp2, MGC81502; NHP2 ribonucleoprotein homolog; K11... 33.1 0.23
tpv:TP04_0176 40S ribosomal protein L7Ae 32.0 0.43
cpv:cgd3_760 HMG-like nuclear protein, Nhp2p, pelota RNA bindi... 32.0 0.44
sce:YDL208W NHP2; Nhp2p; K11129 H/ACA ribonucleoprotein comple... 32.0 0.45
dre:415231 zgc:86738 32.0 0.45
ath:AT5G64840 ATGCN5; ATGCN5 (A. THALIANA GENERAL CONTROL NON-... 30.4 1.4
cpv:cgd7_980 protein with MYND plus SET domains plus 5 Ank rep... 29.6 2.5
ath:AT5G09930 ATGCN2; ATGCN2; transporter 28.1 6.2
> tgo:TGME49_061570 60S ribosomal protein L7a, putative ; K02936
large subunit ribosomal protein L7Ae
Length=276
Score = 169 bits (428), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 78/107 (72%), Positives = 95/107 (88%), Gaps = 0/107 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
P++LVC++P LCRKK+VPYCIVKGK+RLGQLVH+K V+A+D VRKEDQAELEAQCK Y
Sbjct 170 PVELVCFIPQLCRKKEVPYCIVKGKSRLGQLVHQKNCPVLAIDNVRKEDQAELEAQCKIY 229
Query 61 RARFKDNSDLRRRWGGGHLGIKSQHAQEKKQKLIDAEMRKKGGLQIA 107
RA F DNS++RRRWGGG GIKSQHAQ+KK+KLI+ E++KK GLQ+A
Sbjct 230 RAMFNDNSEVRRRWGGGINGIKSQHAQQKKEKLINIELKKKMGLQLA 276
> tpv:TP02_0083 60S ribosomal protein L7a; K02936 large subunit
ribosomal protein L7Ae
Length=266
Score = 140 bits (354), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 64/104 (61%), Positives = 81/104 (77%), Gaps = 0/104 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV WLPALCRKK+VPYCI+K K+RLG+LVH+KT +V+AVD VRKEDQAE + + +
Sbjct 160 PIELVLWLPALCRKKEVPYCIIKSKSRLGKLVHQKTASVVAVDNVRKEDQAEFDNLVRTF 219
Query 61 RARFKDNSDLRRRWGGGHLGIKSQHAQEKKQKLIDAEMRKKGGL 104
A F +NS+LR+RWGG +GIKSQH K++ LI E KK GL
Sbjct 220 TAMFNENSELRKRWGGHIMGIKSQHMIAKREALIAMENAKKLGL 263
> cpv:cgd8_430 60S ribosomal protein L7A ; K02936 large subunit
ribosomal protein L7Ae
Length=263
Score = 139 bits (351), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 59/104 (56%), Positives = 85/104 (81%), Gaps = 0/104 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI++VC+LPALCR+KDV +CI+ GKARLG+L+ KKT AV+A++ VRKEDQAE + C N+
Sbjct 158 PIEIVCFLPALCRRKDVAFCIIGGKARLGKLIGKKTAAVVAIEGVRKEDQAEFDLLCNNF 217
Query 61 RARFKDNSDLRRRWGGGHLGIKSQHAQEKKQKLIDAEMRKKGGL 104
RA++ DN +LRR+WGGG +G+K+QH ++++K++ E KK GL
Sbjct 218 RAQYNDNVELRRKWGGGIMGVKAQHVIKRREKILAMEQAKKVGL 261
> bbo:BBOV_III010580 17.m07914; ribosomal protein L7Ae family
protein; K02936 large subunit ribosomal protein L7Ae
Length=269
Score = 137 bits (346), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 63/106 (59%), Positives = 80/106 (75%), Gaps = 0/106 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
P++LV WLP LCRKK+VPYCI+KGK+RLG+LVH++T +V+AVD +RKEDQAE E K +
Sbjct 163 PLELVMWLPTLCRKKEVPYCIIKGKSRLGKLVHQRTASVVAVDNIRKEDQAEFEGMIKAF 222
Query 61 RARFKDNSDLRRRWGGGHLGIKSQHAQEKKQKLIDAEMRKKGGLQI 106
A F DN ++RRRWGG +GIKSQ + K + LI E KK GL I
Sbjct 223 TAMFNDNVEIRRRWGGHIMGIKSQRQEAKVKALIAMENAKKLGLAI 268
> pfa:PF14_0231 60S ribosomal protein L7-3, putative; K02936 large
subunit ribosomal protein L7Ae
Length=283
Score = 103 bits (258), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 49/92 (53%), Positives = 65/92 (70%), Gaps = 0/92 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV +LPALCR K+VPYCIVK KA LG+LVHKKT + +++V+KEDQ +L+ K
Sbjct 176 PIELVLFLPALCRLKEVPYCIVKDKATLGKLVHKKTATAVCLESVKKEDQEKLDYFAKVC 235
Query 61 RARFKDNSDLRRRWGGGHLGIKSQHAQEKKQK 92
+ F DN DLRR+WGG + KS ++ K K
Sbjct 236 KENFNDNVDLRRKWGGQKMSAKSMLLKKMKDK 267
> ath:AT3G62870 60S ribosomal protein L7A (RPL7aB); K02936 large
subunit ribosomal protein L7Ae
Length=256
Score = 103 bits (256), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 48/98 (48%), Positives = 67/98 (68%), Gaps = 1/98 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV WLPALCRK +VPYCIVKGK+RLG +VH+KT A + + V+ ED+ E +
Sbjct 153 PIELVVWLPALCRKMEVPYCIVKGKSRLGAVVHQKTAAALCLTTVKNEDKLEFSKILEAI 212
Query 61 RARFKDN-SDLRRRWGGGHLGIKSQHAQEKKQKLIDAE 97
+A F D + R++WGGG +G KSQ + K+++I E
Sbjct 213 KANFNDKYEEYRKKWGGGIMGSKSQAKTKAKERVIAKE 250
> ath:AT2G47610 60S ribosomal protein L7A (RPL7aA); K02936 large
subunit ribosomal protein L7Ae
Length=257
Score = 101 bits (252), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 47/98 (47%), Positives = 67/98 (68%), Gaps = 1/98 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV WLPALCRK +VPYCIVKGK+RLG +VH+KT + + + V+ ED+ E +
Sbjct 154 PIELVVWLPALCRKMEVPYCIVKGKSRLGAVVHQKTASCLCLTTVKNEDKLEFSKILEAI 213
Query 61 RARFKDN-SDLRRRWGGGHLGIKSQHAQEKKQKLIDAE 97
+A F D + R++WGGG +G KSQ + K+++I E
Sbjct 214 KANFNDKYEEYRKKWGGGIMGSKSQAKTKAKERVIAKE 251
> sce:YLL045C RPL8B, KRB1, SCL41; Rpl8bp; K02936 large subunit
ribosomal protein L7Ae
Length=256
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 49/97 (50%), Positives = 64/97 (65%), Gaps = 1/97 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV +LPALC+K VPY I+KGKARLG LV++KT AV A+ VR ED+A L
Sbjct 159 PIELVVFLPALCKKMGVPYAIIKGKARLGTLVNQKTSAVAALTEVRAEDEAALAKLVSTI 218
Query 61 RARFKDNSD-LRRRWGGGHLGIKSQHAQEKKQKLIDA 96
A F D D +++ WGGG LG K+Q +K+ K D+
Sbjct 219 DANFADKYDEVKKHWGGGILGNKAQAKMDKRAKTSDS 255
> sce:YHL033C RPL8A, MAK7; Rpl8ap; K02936 large subunit ribosomal
protein L7Ae
Length=256
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 50/97 (51%), Positives = 64/97 (65%), Gaps = 1/97 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV +LPALC+K VPY IVKGKARLG LV++KT AV A+ VR ED+A L
Sbjct 159 PIELVVFLPALCKKMGVPYAIVKGKARLGTLVNQKTSAVAALTEVRAEDEAALAKLVSTI 218
Query 61 RARFKDNSD-LRRRWGGGHLGIKSQHAQEKKQKLIDA 96
A F D D +++ WGGG LG K+Q +K+ K D+
Sbjct 219 DANFADKYDEVKKHWGGGILGNKAQAKMDKRAKNSDS 255
> xla:443912 rpl7a, MGC80199, surf3, trup; ribosomal protein L7a;
K02936 large subunit ribosomal protein L7Ae
Length=266
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 50/87 (57%), Positives = 58/87 (66%), Gaps = 7/87 (8%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAEL----EAQ 56
PI+LV +LPALCRK VPYCIVKGKARLG+LVH+KT +A V ED+ L EA
Sbjct 163 PIELVVFLPALCRKMGVPYCIVKGKARLGRLVHRKTCTSVAFAQVNPEDKGALAKLVEAV 222
Query 57 CKNYRARFKDNSDLRRRWGGGHLGIKS 83
NY RF ++RR WGGG LG KS
Sbjct 223 KTNYNDRF---DEIRRHWGGGILGPKS 246
> hsa:6130 RPL7A, SURF3, TRUP; ribosomal protein L7a; K02936 large
subunit ribosomal protein L7Ae
Length=266
Score = 92.8 bits (229), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 45/84 (53%), Positives = 55/84 (65%), Gaps = 1/84 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV +LPALCRK VPYCI+KGKARLG+LVH+KT +A V ED+ L +
Sbjct 163 PIELVVFLPALCRKMGVPYCIIKGKARLGRLVHRKTCTTVAFTQVNSEDKGALAKLVEAI 222
Query 61 RARFKDNSD-LRRRWGGGHLGIKS 83
R + D D +RR WGG LG KS
Sbjct 223 RTNYNDRYDEIRRHWGGNVLGPKS 246
> xla:100037086 hypothetical protein LOC100037086; K02936 large
subunit ribosomal protein L7Ae
Length=266
Score = 92.8 bits (229), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 45/84 (53%), Positives = 54/84 (64%), Gaps = 1/84 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV +LPALCRK VPYCI+KGKARLG LVH+KT +A V ED+ L +
Sbjct 163 PIELVVFLPALCRKMGVPYCIIKGKARLGHLVHRKTCTTVAFTQVNSEDKGALAKLVEAI 222
Query 61 RARFKDNSD-LRRRWGGGHLGIKS 83
R + D D +RR WGG LG KS
Sbjct 223 RTNYNDRYDEIRRHWGGNVLGPKS 246
> mmu:27176 Rpl7a, MGC103146, Surf3; ribosomal protein L7A; K02936
large subunit ribosomal protein L7Ae
Length=266
Score = 92.8 bits (229), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 45/84 (53%), Positives = 54/84 (64%), Gaps = 1/84 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV +LPALCRK VPYCI+KGKARLG LVH+KT +A V ED+ L +
Sbjct 163 PIELVVFLPALCRKMGVPYCIIKGKARLGHLVHRKTCTTVAFTQVNSEDKGALAKLVEAI 222
Query 61 RARFKDNSD-LRRRWGGGHLGIKS 83
R + D D +RR WGG LG KS
Sbjct 223 RTNYNDRYDEIRRHWGGNVLGPKS 246
> dre:337010 rpl7a, wu:fa95b03, zgc:73183, zgc:86667; ribosomal
protein L7a; K02936 large subunit ribosomal protein L7Ae
Length=266
Score = 92.4 bits (228), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 54/107 (50%), Positives = 65/107 (60%), Gaps = 7/107 (6%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAEL----EAQ 56
PI+LV +LPALCRK VPYCIVKGKARLG+LVH+KT IA ED+A L EA
Sbjct 163 PIELVVFLPALCRKMGVPYCIVKGKARLGRLVHRKTCTSIAFTQTNPEDKAALAKLVEAI 222
Query 57 CKNYRARFKDNSDLRRRWGGGHLGIKSQHAQEKKQKLIDAEMRKKGG 103
NY R++ ++RR WGG LG KS K +K E+ K G
Sbjct 223 KTNYNDRYE---EIRRHWGGNVLGPKSTARIAKLEKAKAKELATKLG 266
> mmu:100504876 60S ribosomal protein L7a-like; K02936 large subunit
ribosomal protein L7Ae
Length=278
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 49/105 (46%), Positives = 60/105 (57%), Gaps = 2/105 (1%)
Query 1 PID-LVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKN 59
PI+ LV +LPALCRK VPYCI+KGKARLG LVH+KT +A V ED+ L +
Sbjct 174 PIEMLVVFLPALCRKMGVPYCIIKGKARLGCLVHRKTCTTVAFTRVNSEDKGALAKLVEA 233
Query 60 YRARFKDNSD-LRRRWGGGHLGIKSQHAQEKKQKLIDAEMRKKGG 103
R + D D + R WGG LG KS K +K E+ K G
Sbjct 234 IRTNYNDRYDEIHRHWGGNVLGPKSVACLAKLEKAKAKELAPKLG 278
> mmu:626683 Gm11362, OTTMUSG00000000657; predicted gene 11362;
K02936 large subunit ribosomal protein L7Ae
Length=266
Score = 82.0 bits (201), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 41/84 (48%), Positives = 52/84 (61%), Gaps = 1/84 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV +LPALC+K VPYCI+KGKARLG LVH+KT +A V E++ L +
Sbjct 163 PIELVVFLPALCQKMGVPYCIIKGKARLGHLVHRKTCTTVAFTQVNSENKGALAKLVEAI 222
Query 61 RARFKDNSD-LRRRWGGGHLGIKS 83
R + D D +R WG LG KS
Sbjct 223 RTNYNDIYDKIRCHWGDNILGPKS 246
> dre:751694 MGC153325; zgc:153325; K02936 large subunit ribosomal
protein L7Ae
Length=269
Score = 80.9 bits (198), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 46/105 (43%), Positives = 64/105 (60%), Gaps = 7/105 (6%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAEL----EAQ 56
P++LV +LP+LCRK VPYCIVK K+RLG+LV +KT + +A+ V D+ L E
Sbjct 165 PVELVIFLPSLCRKMGVPYCIVKDKSRLGRLVRRKTCSAVALTQVEAGDRNALAKLIETV 224
Query 57 CKNYRARFKDNSDLRRRWGGGHLGIKSQHAQEKKQKLIDAEMRKK 101
NY RF+ ++++ WGGG LG KS K +K EM +K
Sbjct 225 KTNYNDRFE---EIKKHWGGGTLGSKSNARIAKIEKAKAKEMAQK 266
> cel:Y24D9A.4 rpl-7A; Ribosomal Protein, Large subunit family
member (rpl-7A); K02936 large subunit ribosomal protein L7Ae
Length=245
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 43/105 (40%), Positives = 62/105 (59%), Gaps = 4/105 (3%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
P+++V LPALCRK +VPY I+KGKA LG +V +KT A +A+ V ED++ L +
Sbjct 144 PLEIVLHLPALCRKYNVPYAIIKGKASLGTVVRRKTTAAVALVDVNPEDKSALNKLVETV 203
Query 61 RARFKD-NSDLRRRWGGGHLGIKSQHAQEKKQKLIDAEMRKKGGL 104
F + + ++R+ WGGG + KS KK K+ A R G L
Sbjct 204 NNNFSERHEEIRKHWGGGVMSAKS---DAKKLKIERARARDLGKL 245
> pfa:PFD0960c 60S ribosomal protein L7Ae/L30e, putative
Length=236
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQL 31
PI+++C LP+ C + +PY V K +L L
Sbjct 148 PINIICHLPSFCEQYKIPYTFVTTKNKLAHL 178
> cel:Y48A6B.3 hypothetical protein; K11129 H/ACA ribonucleoprotein
complex subunit 2
Length=163
Score = 36.2 bits (82), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 26/43 (60%), Gaps = 1/43 (2%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLV-HKKTGAVIAV 42
PID+ +P +C +K++PY + + +LG V H++ +I V
Sbjct 96 PIDVYSHIPGICEEKEIPYVYIPSREQLGLAVGHRRPSILIFV 138
> dre:321114 nhp2l1, Ssfa1, cb127, sb:cb127, zgc:56265; NHP2 non-histone
chromosome protein 2-like 1 (S. cerevisiae); K12845
U4/U6 small nuclear ribonucleoprotein SNU13
Length=128
Score = 36.2 bits (82), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLG 29
P++++ LP LC K+VPY V+ K LG
Sbjct 62 PLEIILHLPLLCEDKNVPYVFVRSKQALG 90
> dre:393282 MGC56066, MGC77417, zgc:77417; zgc:56066; K12845
U4/U6 small nuclear ribonucleoprotein SNU13
Length=128
Score = 36.2 bits (82), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLG 29
P++++ LP LC K+VPY V+ K LG
Sbjct 62 PLEIILHLPLLCEDKNVPYVFVRSKQALG 90
> tpv:TP04_0892 hypothetical protein; K12845 U4/U6 small nuclear
ribonucleoprotein SNU13
Length=129
Score = 35.8 bits (81), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQ 30
P++++ LP LC K+VPY V K LG+
Sbjct 63 PLEIILHLPLLCEDKNVPYIFVHSKVALGR 92
> xla:495253 nhp2l1-a, fa-1, fa1, hoip, nhp2l1, nhpx, otk27, snrnp15-5,
snu13, spag12, ssfa1; NHP2 non-histone chromosome
protein 2-like 1; K12845 U4/U6 small nuclear ribonucleoprotein
SNU13
Length=128
Score = 35.4 bits (80), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLG 29
P++++ LP LC K+VPY V+ K LG
Sbjct 62 PLEIILHLPLLCEDKNVPYVFVRSKQALG 90
> xla:380416 nhp2l1-b, MGC53430, fa-1, fa1, hoip, nhpx, otk27,
snrnp15-5, snu13, spag12, ssfa1; NHP2 non-histone chromosome
protein 2-like 1; K12845 U4/U6 small nuclear ribonucleoprotein
SNU13
Length=128
Score = 35.4 bits (80), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLG 29
P++++ LP LC K+VPY V+ K LG
Sbjct 62 PLEIILHLPLLCEDKNVPYVFVRSKQALG 90
> dre:100003787 nhp2, MGC73227, nola2, wu:fb36b10, wu:fd61a03,
zgc:73227; NHP2 ribonucleoprotein homolog (yeast); K11129 H/ACA
ribonucleoprotein complex subunit 2
Length=150
Score = 35.4 bits (80), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQ 30
PID+ C LP +C + +PY V K LG
Sbjct 84 PIDVYCHLPIMCEDRSLPYAYVPSKVDLGS 113
> mmu:20826 Nhp2l1, FA-1, Fta1, Ssfa1; NHP2 non-histone chromosome
protein 2-like 1 (S. cerevisiae); K12845 U4/U6 small nuclear
ribonucleoprotein SNU13
Length=128
Score = 35.0 bits (79), Expect = 0.052, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQ 30
P++++ LP LC K+VPY V+ K LG+
Sbjct 62 PLEIILHLPLLCEDKNVPYVFVRSKQALGR 91
> hsa:4809 NHP2L1, 15.5K, FA-1, FA1, NHPX, OTK27, SNRNP15-5, SNU13,
SPAG12, SSFA1; NHP2 non-histone chromosome protein 2-like
1 (S. cerevisiae); K12845 U4/U6 small nuclear ribonucleoprotein
SNU13
Length=128
Score = 35.0 bits (79), Expect = 0.052, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQ 30
P++++ LP LC K+VPY V+ K LG+
Sbjct 62 PLEIILHLPLLCEDKNVPYVFVRSKQALGR 91
> mmu:434401 Gm5616, EG434401; predicted gene 5616; K12845 U4/U6
small nuclear ribonucleoprotein SNU13
Length=128
Score = 35.0 bits (79), Expect = 0.053, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQ 30
P++++ LP LC K+VPY V+ K LG+
Sbjct 62 PLEIILHLPLLCEDKNVPYVFVRSKQALGR 91
> mmu:100045968 NHP2-like protein 1-like; K12845 U4/U6 small nuclear
ribonucleoprotein SNU13
Length=134
Score = 35.0 bits (79), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQ 30
P++++ LP LC K+VPY V+ K LG+
Sbjct 62 PLEIILHLPLLCEDKNVPYVFVRSKQALGR 91
> mmu:100505330 NHP2-like protein 1-like; K12845 U4/U6 small nuclear
ribonucleoprotein SNU13
Length=128
Score = 35.0 bits (79), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQ 30
P++++ LP LC K+VPY V+ K LG+
Sbjct 62 PLEIILHLPLLCEDKNVPYVFVRSKQALGR 91
> bbo:BBOV_III009740 17.m07845; ribosomal protein L7A; K12845
U4/U6 small nuclear ribonucleoprotein SNU13
Length=128
Score = 35.0 bits (79), Expect = 0.065, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQ 30
P++++ LP +C K++PY VK K LG+
Sbjct 62 PLEIILHLPLVCEDKNIPYIFVKSKIALGR 91
> mmu:52530 Nhp2, 2410130M07Rik, D11Ertd175e, Nola2; NHP2 ribonucleoprotein
homolog (yeast); K11129 H/ACA ribonucleoprotein
complex subunit 2
Length=153
Score = 34.7 bits (78), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 23/43 (53%), Gaps = 1/43 (2%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLV-HKKTGAVIAV 42
PI++ C LP LC +++PY + K LG K+ VI V
Sbjct 87 PIEVYCHLPVLCEDQNLPYVYIPSKTDLGAATGSKRPTCVIMV 129
> sce:YEL026W SNU13; RNA binding protein, part of U3 snoRNP involved
in rRNA processing, part of U4/U6-U5 tri-snRNP involved
in mRNA splicing, similar to human 15.5K protein; K12845
U4/U6 small nuclear ribonucleoprotein SNU13
Length=126
Score = 34.7 bits (78), Expect = 0.077, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQ 30
PI+++ LP LC K+VPY V + LG+
Sbjct 60 PIEILLHLPLLCEDKNVPYVFVPSRVALGR 89
> pfa:PF11_0250 high mobility group-like protein NHP2, putative;
K12845 U4/U6 small nuclear ribonucleoprotein SNU13
Length=145
Score = 34.3 bits (77), Expect = 0.089, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 32/61 (52%), Gaps = 2/61 (3%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
P++++ +P +C K+ PY V+ K LG+ + +VIA V K D + LE Q
Sbjct 79 PLEIISHIPLVCEDKNTPYVYVRSKMALGRAC-GISRSVIATSIVTK-DGSPLETQITEL 136
Query 61 R 61
+
Sbjct 137 K 137
> tgo:TGME49_113560 60S ribosomal protein L7a, putative ; K11129
H/ACA ribonucleoprotein complex subunit 2
Length=173
Score = 33.9 bits (76), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 30/57 (52%), Gaps = 3/57 (5%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLV-HKKTGAVIAVDAVRKEDQAELEAQ 56
PI+++ LP LC +KDV Y + K LG K+ +VI + ED E++ +
Sbjct 95 PIEIIAHLPILCEEKDVVYAYLCSKKTLGHAFRSKRPASVIMITP--GEDMPEVDGE 149
> cpv:cgd6_3850 HOI-POLLOI protein; U4/U6.U5 snRNP component;
Snu13p; pelota RNA binding domain containing protein ; K12845
U4/U6 small nuclear ribonucleoprotein SNU13
Length=134
Score = 33.5 bits (75), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 29/56 (51%), Gaps = 2/56 (3%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQ 56
P++++ LP +C K+ PY V+ K LG+ + A A+ +D + L +Q
Sbjct 68 PLEILLHLPLVCEDKNTPYVFVRSKVALGRACGVSRPVIAA--AITSKDGSSLSSQ 121
> ath:AT5G08180 ribosomal protein L7Ae/L30e/S12e/Gadd45 family
protein; K11129 H/ACA ribonucleoprotein complex subunit 2
Length=156
Score = 33.1 bits (74), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQ 30
PID++ LP LC + VPY V K L Q
Sbjct 78 PIDVITHLPILCEEAGVPYVYVPSKEDLAQ 107
> xla:414481 nhp2, MGC81502; NHP2 ribonucleoprotein homolog; K11129
H/ACA ribonucleoprotein complex subunit 2
Length=149
Score = 33.1 bits (74), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 28/58 (48%), Gaps = 3/58 (5%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLV-HKKTGAVIAVDAVRKEDQAELEAQC 57
PI++ C +P +C + +PY V K+ LG K+ VI + ED E +C
Sbjct 83 PIEVYCHIPVMCEDRGIPYSYVPSKSDLGAAAGSKRPTCVILIKP--HEDYQEAYDEC 138
> tpv:TP04_0176 40S ribosomal protein L7Ae
Length=177
Score = 32.0 bits (71), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 25/48 (52%), Gaps = 2/48 (4%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKK--TGAVIAVDAVR 46
P+D+V +P LC + + Y V K L + H K T AV+ V ++
Sbjct 94 PVDVVSHVPVLCEELSISYAYVASKRVLSDVCHSKRPTCAVLVVKPLK 141
> cpv:cgd3_760 HMG-like nuclear protein, Nhp2p, pelota RNA binding
domain containing protein ; K11129 H/ACA ribonucleoprotein
complex subunit 2
Length=172
Score = 32.0 bits (71), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 13/52 (25%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAE 52
P+D++ +P LC +K++ Y + K LG + K A + + + E +
Sbjct 99 PVDIIAHIPILCEEKNIYYGYLGSKKTLGTICKSKRPASVLMISFNSESSVQ 150
> sce:YDL208W NHP2; Nhp2p; K11129 H/ACA ribonucleoprotein complex
subunit 2
Length=156
Score = 32.0 bits (71), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 15/29 (51%), Gaps = 0/29 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLG 29
P D++ +P LC VPY + K LG
Sbjct 83 PADVISHIPVLCEDHSVPYIFIPSKQDLG 111
> dre:415231 zgc:86738
Length=257
Score = 32.0 bits (71), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 20/40 (50%), Gaps = 1/40 (2%)
Query 6 CWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAV 45
CW LC IV G R QL+HK T +V+ ++ +
Sbjct 77 CWFSLLCSAASF-IVIVIGLLRYAQLIHKHTNSVLNIEGL 115
> ath:AT5G64840 ATGCN5; ATGCN5 (A. THALIANA GENERAL CONTROL NON-REPRESSIBLE
5); transporter
Length=692
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 29/112 (25%), Positives = 51/112 (45%), Gaps = 12/112 (10%)
Query 2 IDLVCWLPALCRKKDVPYCIVK-GKARLGQLVHKKTGAVIAVDAVRKEDQAE-------- 52
+D + WL +K+DVP I+ +A L QL K + V + + ++
Sbjct 292 LDTIEWLEGYLQKQDVPMVIISHDRAFLDQLCTKIVETEMGVSRTFEGNYSQYVISKAEW 351
Query 53 LEAQCKNYRARFKD---NSDLRRRWGGGHLGIKSQHAQEKKQKLIDAEMRKK 101
+E Q + + KD DL R G G ++ A++K +KL + E+ +K
Sbjct 352 IETQNAAWEKQQKDIDSTKDLIARLGAGANSGRASTAEKKLEKLQEQELIEK 403
> cpv:cgd7_980 protein with MYND plus SET domains plus 5 Ank repeats
Length=1560
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 18/56 (32%), Positives = 28/56 (50%), Gaps = 3/56 (5%)
Query 35 KTGAVIAVDAVRKEDQAELEAQCKNYRARFKDNSDLRRRWGGGHLGIKSQHAQEKK 90
K A+IA D +RK ++E K Y+ K+NS+ G + I+ +HA K
Sbjct 903 KKSAIIARDMIRKAIALDMEKSSKTYK---KENSEDNGEHLGANACIRKKHAFASK 955
> ath:AT5G09930 ATGCN2; ATGCN2; transporter
Length=678
Score = 28.1 bits (61), Expect = 6.2, Method: Composition-based stats.
Identities = 32/112 (28%), Positives = 51/112 (45%), Gaps = 12/112 (10%)
Query 2 IDLVCWLPALCRKKDVPYCIVK-GKARLGQL----VHKKTGAVIAVDAVRKE---DQAEL 53
+D + WL K+DVP I+ +A L QL V + G D + +AEL
Sbjct 278 LDTIEWLEGYLIKQDVPMVIISHDRAFLDQLCTKIVETEMGVSRTFDGNYSQYVISKAEL 337
Query 54 -EAQCKNYRARFKD---NSDLRRRWGGGHLGIKSQHAQEKKQKLIDAEMRKK 101
EAQ + + K+ DL R G ++ A++K +KL + E+ +K
Sbjct 338 VEAQYAAWEKQQKEIEATKDLISRLSAGANSGRASSAEKKLEKLQEEELIEK 389
Lambda K H
0.321 0.137 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2007980300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40