bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3861_orf2
Length=144
Score E
Sequences producing significant alignments: (Bits) Value
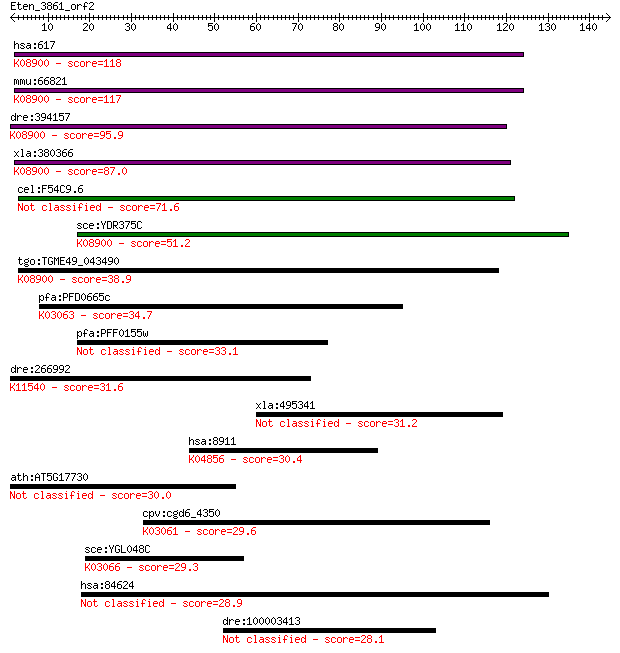
hsa:617 BCS1L, BCS, BCS1, BJS, FLNMS, GRACILE, Hs.6719, PTD, h... 118 6e-27
mmu:66821 Bcs1l, 9130022O19Rik; BCS1-like (yeast); K08900 mito... 117 2e-26
dre:394157 bcs1l, MGC56205, zgc:56205; BCS1-like (yeast); K089... 95.9 4e-20
xla:380366 bcs1l, MGC53156; BCS1-like (S. cerevisiae); K08900 ... 87.0 2e-17
cel:F54C9.6 bcs-1; BCS1 (mitochondrial chaperone) homolog fami... 71.6 8e-13
sce:YDR375C BCS1; Bcs1p; K08900 mitochondrial chaperone BCS1 51.2 1e-06
tgo:TGME49_043490 bcs1 protein, putative (EC:3.6.4.6); K08900 ... 38.9 0.005
pfa:PFD0665c 26S proteasome AAA-ATPase subunit RPT3, putative ... 34.7 0.10
pfa:PFF0155w Bcs1 protein, putative 33.1 0.29
dre:266992 cad, cb456, si:dkey-221h15.3, wu:fc30c12, wu:fc33d0... 31.6 0.98
xla:495341 mtmr10-a, mtmr10; myotubularin related protein 10 31.2
hsa:8911 CACNA1I, Cav3.3, KIAA1120; calcium channel, voltage-d... 30.4 2.0
ath:AT5G17730 AAA-type ATPase family protein 30.0 2.4
cpv:cgd6_4350 26S proteasome regulatory subunit 7 (RPT1)-like.... 29.6 3.8
sce:YGL048C RPT6, CIM3, CRL3, SCB68, SUG1; One of six ATPases ... 29.3 4.2
hsa:84624 FNDC1, AGS8, FNDC2, KIAA1866, MEL4B3, bA243O10.1, dJ... 28.9 6.5
dre:100003413 Jmjd2al protein-like 28.1 9.7
> hsa:617 BCS1L, BCS, BCS1, BJS, FLNMS, GRACILE, Hs.6719, PTD,
h-BCS; BCS1-like (S. cerevisiae); K08900 mitochondrial chaperone
BCS1
Length=419
Score = 118 bits (296), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 60/122 (49%), Positives = 73/122 (59%), Gaps = 0/122 (0%)
Query 2 ENPKKYRGLARLPLRGALNPRGGGGSPKARIVFMPPPQMERLDPALKPPGGVDKKEYGGP 61
ENP KY+GL RL G LN G S +ARIVFM ++RLDPAL PG VD KEY G
Sbjct 296 ENPVKYQGLGRLTFSGLLNALDGVASTEARIVFMTTNHVDRLDPALIRPGRVDLKEYVGY 355
Query 62 PPRGKVPQIFQGFFPGQATSLVENFENRVLQTPPQIIPPQGRGSFLRKKKTPPGALQKTK 121
++ Q+FQ F+PGQA SL ENF VL+ QI P Q +G F+ K P GA+ +
Sbjct 356 CSHWQLTQMFQRFYPGQAPSLAENFAEHVLRATNQISPAQVQGYFMLYKNDPVGAIHNAE 415
Query 122 FL 123
L
Sbjct 416 SL 417
> mmu:66821 Bcs1l, 9130022O19Rik; BCS1-like (yeast); K08900 mitochondrial
chaperone BCS1
Length=418
Score = 117 bits (292), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 59/122 (48%), Positives = 73/122 (59%), Gaps = 0/122 (0%)
Query 2 ENPKKYRGLARLPLRGALNPRGGGGSPKARIVFMPPPQMERLDPALKPPGGVDKKEYGGP 61
ENP KY+GL RL G LN G S +ARIVFM ++RLDPAL PG VD KEY G
Sbjct 296 ENPIKYQGLGRLTFSGLLNALDGVASTEARIVFMTTNYIDRLDPALIRPGRVDLKEYVGY 355
Query 62 PPRGKVPQIFQGFFPGQATSLVENFENRVLQTPPQIIPPQGRGSFLRKKKTPPGALQKTK 121
++ Q+FQ F+PGQA SL ENF VL+ +I P Q +G F+ K P GA+ +
Sbjct 356 CSHWQLTQMFQRFYPGQAPSLAENFAEHVLKATSEISPAQVQGYFMLYKNDPMGAVHNIE 415
Query 122 FL 123
L
Sbjct 416 SL 417
> dre:394157 bcs1l, MGC56205, zgc:56205; BCS1-like (yeast); K08900
mitochondrial chaperone BCS1
Length=420
Score = 95.9 bits (237), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 45/119 (37%), Positives = 66/119 (55%), Gaps = 0/119 (0%)
Query 1 PENPKKYRGLARLPLRGALNPRGGGGSPKARIVFMPPPQMERLDPALKPPGGVDKKEYGG 60
ENP Y+G+ RL G LN G S +ARIVFM +ERLDPAL PG VD K+Y G
Sbjct 296 TENPLAYQGMGRLTFSGLLNALDGVASSEARIVFMTTNFIERLDPALVRPGRVDLKQYVG 355
Query 61 PPPRGKVPQIFQGFFPGQATSLVENFENRVLQTPPQIIPPQGRGSFLRKKKTPPGALQK 119
++ Q+F+ F+P ++ + ++F + L + Q +G F+ K P GA++
Sbjct 356 HCSHWQLTQMFRRFYPQESAAEADHFSEQALAAHTDLSAAQVQGHFMLYKTDPAGAIKN 414
> xla:380366 bcs1l, MGC53156; BCS1-like (S. cerevisiae); K08900
mitochondrial chaperone BCS1
Length=419
Score = 87.0 bits (214), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 47/119 (39%), Positives = 64/119 (53%), Gaps = 0/119 (0%)
Query 2 ENPKKYRGLARLPLRGALNPRGGGGSPKARIVFMPPPQMERLDPALKPPGGVDKKEYGGP 61
+NP Y+G+ RL G LN G S +ARIVFM ++RLDPAL PG VD K+Y G
Sbjct 296 QNPTAYQGMGRLTFSGLLNALDGVASTEARIVFMTTNHIDRLDPALIRPGRVDVKQYVGH 355
Query 62 PPRGKVPQIFQGFFPGQATSLVENFENRVLQTPPQIIPPQGRGSFLRKKKTPPGALQKT 120
++ Q+F F+P Q E F + L + +I Q +G F+ K P GA+Q
Sbjct 356 CTNWQLSQMFLRFYPDQTAGQSEAFASAALSSSDKISAAQVQGHFMMHKTDPDGAIQNV 414
> cel:F54C9.6 bcs-1; BCS1 (mitochondrial chaperone) homolog family
member (bcs-1)
Length=442
Score = 71.6 bits (174), Expect = 8e-13, Method: Composition-based stats.
Identities = 43/120 (35%), Positives = 62/120 (51%), Gaps = 1/120 (0%)
Query 3 NPKKYRGLARLPLRGALNPRGGGGSPKARIVFMPPPQMERLDPALKPPGGVDKKEYGGPP 62
N Y+GL+R+ G LN G + R+ FM +ERLDPAL PG VD+K+Y G
Sbjct 317 NHPAYQGLSRVTFSGLLNALDGVACAEERLTFMTTNYVERLDPALIRPGRVDRKQYFGNA 376
Query 63 PRGKVPQIFQGFFPGQATS-LVENFENRVLQTPPQIIPPQGRGSFLRKKKTPPGALQKTK 121
G + ++F F+ + S L + F RV + ++ P +G FL K+ P AL K
Sbjct 377 TDGMLSKMFSRFYRQPSDSVLADEFVKRVSEHKTELSPAMIQGHFLMYKQDPRAALDNIK 436
> sce:YDR375C BCS1; Bcs1p; K08900 mitochondrial chaperone BCS1
Length=456
Score = 51.2 bits (121), Expect = 1e-06, Method: Composition-based stats.
Identities = 37/118 (31%), Positives = 55/118 (46%), Gaps = 5/118 (4%)
Query 17 GALNPRGGGGSPKARIVFMPPPQMERLDPALKPPGGVDKKEYGGPPPRGKVPQIFQGFFP 76
G LN G S + I FM E+LD A+ PG +D K + G +V ++F F+P
Sbjct 344 GLLNALDGVTSSEETITFMTTNHPEKLDAAIMRPGRIDYKVFVGNATPYQVEKMFMKFYP 403
Query 77 GQATSLVENFENRVLQTPPQIIPPQGRGSFLRKKKTPPGALQKTKFLGGGQRGWPHLF 134
G+ T + + F N V + + Q +G F+ K P AL+ L R H+F
Sbjct 404 GE-TDICKKFVNSVKELDITVSTAQLQGLFVMNKDAPHDALKMVSSL----RNANHIF 456
> tgo:TGME49_043490 bcs1 protein, putative (EC:3.6.4.6); K08900
mitochondrial chaperone BCS1
Length=570
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 35/115 (30%), Positives = 51/115 (44%), Gaps = 6/115 (5%)
Query 3 NPKKYRGLARLPLRGALNPRGGGGSPKARIVFMPPPQMERLDPALKPPGGVDKKEYGGPP 62
NP RG+ G LN G + + R+ M ERL +L PG VD K G
Sbjct 393 NPYGMRGV---TFSGLLNALDGIVATEERVTIMTTNHPERLPDSLIRPGRVDIKVRIGYA 449
Query 63 PRGKVPQIFQGFFPGQATSLVENFENRVLQTPPQIIPPQGRGSFLRKKKTPPGAL 117
R ++ + F FFPG+ + + FE + + Q+ + +G FL K AL
Sbjct 450 TRPQLRRQFLRFFPGEDAA-ADKFE--AIMSGIQLSMAELQGFFLFCKDNVDQAL 501
> pfa:PFD0665c 26S proteasome AAA-ATPase subunit RPT3, putative
(EC:3.6.4.8); K03063 26S proteasome regulatory subunit T3
Length=392
Score = 34.7 bits (78), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 37/87 (42%), Gaps = 1/87 (1%)
Query 8 RGLARLPLRGALNPRGGGGSPKARIVFMPPPQMERLDPALKPPGGVDKKEYGGPPPRGKV 67
R + R+ L LN G V M + + LDPAL PG +D+K P R +
Sbjct 256 REVQRILLE-LLNQMDGFDKSTNVKVIMATNRADTLDPALLRPGRLDRKIEFPLPDRKQK 314
Query 68 PQIFQGFFPGQATSLVENFENRVLQTP 94
IFQ S N E+ V++T
Sbjct 315 RLIFQTIISKMNVSSDVNIESFVVRTD 341
> pfa:PFF0155w Bcs1 protein, putative
Length=471
Score = 33.1 bits (74), Expect = 0.29, Method: Composition-based stats.
Identities = 21/60 (35%), Positives = 29/60 (48%), Gaps = 0/60 (0%)
Query 17 GALNPRGGGGSPKARIVFMPPPQMERLDPALKPPGGVDKKEYGGPPPRGKVPQIFQGFFP 76
G LN G + + RI+FM +E+L P L PG VD K + ++F FFP
Sbjct 356 GLLNALDGIVATEERIIFMTTNNIEKLPPTLIRPGRVDMKILIPYANIYQYKKMFLRFFP 415
> dre:266992 cad, cb456, si:dkey-221h15.3, wu:fc30c12, wu:fc33d01,
wu:fc67g02; carbamoyl-phosphate synthetase 2, aspartate
transcarbamylase, and dihydroorotase (EC:2.1.3.2); K11540 carbamoyl-phosphate
synthase / aspartate carbamoyltransferase
/ dihydroorotase [EC:6.3.5.5 2.1.3.2 3.5.2.3]
Length=2230
Score = 31.6 bits (70), Expect = 0.98, Method: Composition-based stats.
Identities = 23/75 (30%), Positives = 36/75 (48%), Gaps = 8/75 (10%)
Query 1 PENPKKYRGLARLPLRGALNPRGGGGSPKARIVFMPPPQMERL-DPALKPPGGVDKKEYG 59
PE P++ + + R A +PR G + F+ PP++ R DP L P + + Y
Sbjct 1842 PERPRQAAPVDAVRSR-APSPRRSAGDGR----FILPPRIHRSSDPGLPPELALPAEAYA 1896
Query 60 GPPPRGKV--PQIFQ 72
PPP ++ PQ Q
Sbjct 1897 HPPPLARILSPQAGQ 1911
> xla:495341 mtmr10-a, mtmr10; myotubularin related protein 10
Length=765
Score = 31.2 bits (69), Expect = 1.2, Method: Composition-based stats.
Identities = 20/59 (33%), Positives = 25/59 (42%), Gaps = 0/59 (0%)
Query 60 GPPPRGKVPQIFQGFFPGQATSLVENFENRVLQTPPQIIPPQGRGSFLRKKKTPPGALQ 118
G PP K Q F + SL+ + +LQ P IIP G +LR T P L
Sbjct 573 GMPPSVKNGMAIQQDFMPRRNSLILKLKPEILQQVPVIIPSSGFEQYLRDWFTKPADLH 631
> hsa:8911 CACNA1I, Cav3.3, KIAA1120; calcium channel, voltage-dependent,
T type, alpha 1I subunit; K04856 voltage-dependent
calcium channel T type alpha-1I
Length=2188
Score = 30.4 bits (67), Expect = 2.0, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 23/45 (51%), Gaps = 0/45 (0%)
Query 44 DPALKPPGGVDKKEYGGPPPRGKVPQIFQGFFPGQATSLVENFEN 88
DP PPG D K PP +V + + FFP +T++ + EN
Sbjct 1841 DPTACPPGRKDSKGELDPPEPMRVGDLGECFFPLSSTAVSPDPEN 1885
> ath:AT5G17730 AAA-type ATPase family protein
Length=470
Score = 30.0 bits (66), Expect = 2.4, Method: Composition-based stats.
Identities = 21/56 (37%), Positives = 25/56 (44%), Gaps = 2/56 (3%)
Query 1 PENPKKYRGLARLPLRGALNPRGGGGSP--KARIVFMPPPQMERLDPALKPPGGVD 54
P+ +G + L L G L G S RIV ERLDPAL PG +D
Sbjct 317 PKTQDDTKGSSMLTLSGLLTCIDGLWSSCGDERIVIFTTTHKERLDPALLRPGRMD 372
> cpv:cgd6_4350 26S proteasome regulatory subunit 7 (RPT1)-like.
AAA atpase ; K03061 26S proteasome regulatory subunit T1
Length=432
Score = 29.6 bits (65), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 39/84 (46%), Gaps = 8/84 (9%)
Query 33 VFMPPPQMERLDPALKPPGGVDKK-EYGGPPPRGKVPQIFQGFFPGQATSLVENFENRVL 91
V M + + LDPAL PG +D+K E+G P G+ QIF+ + S+ + +L
Sbjct 316 VLMATNRPDTLDPALLRPGRLDRKVEFGLPDLEGR-TQIFR--IHAKVMSMERDIRFELL 372
Query 92 QTPPQIIPPQGRGSFLRKKKTPPG 115
+ P G+ +R T G
Sbjct 373 SR----LCPNCTGADIRSVCTEAG 392
> sce:YGL048C RPT6, CIM3, CRL3, SCB68, SUG1; One of six ATPases
of the 19S regulatory particle of the 26S proteasome involved
in the degradation of ubiquitinated substrates; bound by
ubiquitin-protein ligases Ubr1p and Ufd4p; localized mainly
to the nucleus throughout the cell cycle; K03066 26S proteasome
regulatory subunit T6
Length=405
Score = 29.3 bits (64), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 19 LNPRGGGGSPKARIVFMPPPQMERLDPALKPPGGVDKK 56
LN G + K + M +++ LDPAL PG +D+K
Sbjct 276 LNQLDGFETSKNIKIIMATNRLDILDPALLRPGRIDRK 313
> hsa:84624 FNDC1, AGS8, FNDC2, KIAA1866, MEL4B3, bA243O10.1,
dJ322A24.1; fibronectin type III domain containing 1
Length=1894
Score = 28.9 bits (63), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 47/119 (39%), Gaps = 15/119 (12%)
Query 18 ALNPRGGGGSPKARIVFMPPPQMERLDPALKPPGGVDKKEYGGPPPRG-------KVPQI 70
A N RG G KA IV MP ++ + G +K E P PR VP
Sbjct 437 ATNRRGLGPHSKAFIVAMPTTSKADVEQNTEDNGKPEKPEPSSPSPRAPASSQHPSVPAS 496
Query 71 FQGFFPGQATSLVENFENRVLQTPPQIIPPQGRGSFLRKKKTPPGALQKTKFLGGGQRG 129
QG A L+ + +N++L PQ LR KK LQ T+ G + G
Sbjct 497 PQGR---NAKDLLLDLKNKILANGGAPRKPQ-----LRAKKAEELDLQSTEITGEEELG 547
> dre:100003413 Jmjd2al protein-like
Length=2012
Score = 28.1 bits (61), Expect = 9.7, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 25/52 (48%), Gaps = 1/52 (1%)
Query 52 GVDKKEYGGPPPRGK-VPQIFQGFFPGQATSLVENFENRVLQTPPQIIPPQG 102
G K Y PP GK + ++ +GFFPG A S +++ P I+ G
Sbjct 206 GEPKSWYVVPPEHGKRLERLAKGFFPGSAQSCEAFLRHKMTLISPSILRKYG 257
Lambda K H
0.317 0.145 0.465
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2814663556
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40