bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3864_orf1
Length=174
Score E
Sequences producing significant alignments: (Bits) Value
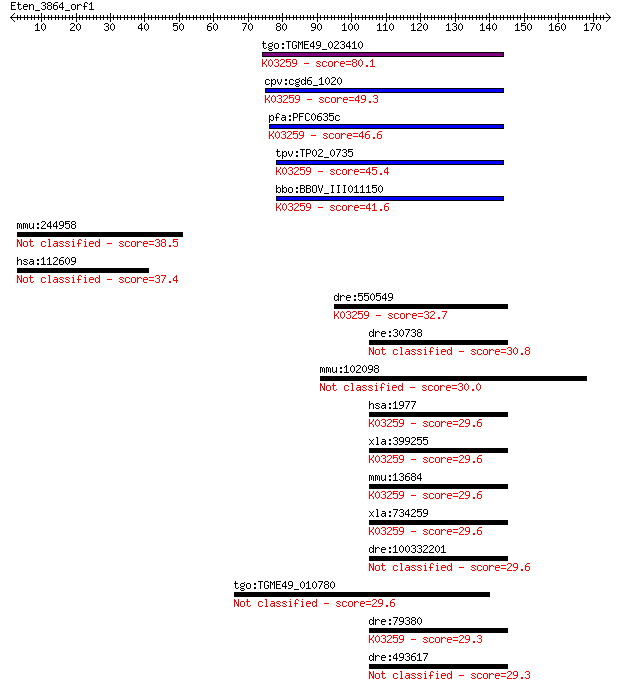
tgo:TGME49_023410 eukaryotic translation initiation factor 4E,... 80.1 3e-15
cpv:cgd6_1020 translation initiation factor if-4E ; K03259 tra... 49.3 6e-06
pfa:PFC0635c translation initiation factor E4, putative; K0325... 46.6 4e-05
tpv:TP02_0735 translation initiation factor E4; K03259 transla... 45.4 9e-05
bbo:BBOV_III011150 17.m10646; translation initiation factor E4... 41.6 0.001
mmu:244958 Mrap2, BB633055; melanocortin 2 receptor accessory ... 38.5 0.011
hsa:112609 MRAP2, C6orf117, RP11-51G5.2, bA51G5.2; melanocorti... 37.4 0.027
dre:550549 eif4e1c, eif4E-1C, fb57e09, wu:fb57e09, zgc:110154;... 32.7 0.56
dre:30738 eif4e1b, Z4ES, eif4e, zeIF4E; eukaryotic translation... 30.8 2.4
mmu:102098 Arhgef18, AI467246, D030053O22Rik; rho/rac guanine ... 30.0 4.7
hsa:1977 EIF4E, CBP, EIF4E1, EIF4EL1, EIF4F, MGC111573; eukary... 29.6 4.8
xla:399255 eif4e, MGC84530; eIF-4E protein; K03259 translation... 29.6 5.4
mmu:13684 Eif4e, EG668879, Eif4e-ps, If4e, MGC103177, eIF-4E; ... 29.6 5.5
xla:734259 eif4e, MGC85107, eIF-4E, eif4e1; eukaryotic transla... 29.6 5.6
dre:100332201 eukaryotic translation initiation factor 4E-1A-like 29.6 5.8
tgo:TGME49_010780 ubiquitin carboxyl-terminal hydrolase, putat... 29.6 6.1
dre:79380 eif4e, eif4e-1, eif4e1a, zgc:86680; eukaryotic trans... 29.3 6.1
dre:493617 zgc:101581 29.3 7.9
> tgo:TGME49_023410 eukaryotic translation initiation factor 4E,
putative ; K03259 translation initiation factor 4E
Length=198
Score = 80.1 bits (196), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 36/70 (51%), Positives = 52/70 (74%), Gaps = 4/70 (5%)
Query 74 STKYLSFNPSVADAVNLDECITEERPADAPMPLQHRWHVWEQIQREAAAADRAADYSQNT 133
+ K+LSFN + +AV+L + + EE D P+PL++ WHVWEQ+Q++ DR+ +YS NT
Sbjct 2 AAKFLSFNQELNEAVDLKDWVKEEPVPDEPLPLRYVWHVWEQVQQD----DRSKEYSDNT 57
Query 134 RDLASFDTVQ 143
RDLA+FDTVQ
Sbjct 58 RDLAAFDTVQ 67
> cpv:cgd6_1020 translation initiation factor if-4E ; K03259 translation
initiation factor 4E
Length=240
Score = 49.3 bits (116), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 40/73 (54%), Gaps = 7/73 (9%)
Query 75 TKYLSFNPSVADAVNLDECITE-ERPADA---PMPLQHRWHVWEQIQREAAAADRAADYS 130
+KYLSFN S+ + L +E + P D P+PL H W VWEQ+ E + DYS
Sbjct 8 SKYLSFNESIEPNLTLPTACSEVDLPEDLISRPLPLSHEWIVWEQLNVETR---KDLDYS 64
Query 131 QNTRDLASFDTVQ 143
T+ +A F +VQ
Sbjct 65 NATKPVARFSSVQ 77
> pfa:PFC0635c translation initiation factor E4, putative; K03259
translation initiation factor 4E
Length=227
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 39/69 (56%), Gaps = 3/69 (4%)
Query 76 KYLSFNPSVADAVNLDECITEER-PADAPMPLQHRWHVWEQIQREAAAADRAADYSQNTR 134
KYL+FN + DA++L E + + P+ LQ+ W +WEQ+ ++ +Y TR
Sbjct 2 KYLTFNKNNRDAIDLSEKLEATKIDLSNPLLLQYNWVIWEQVSDNKIK--QSNNYKDYTR 59
Query 135 DLASFDTVQ 143
LA F++VQ
Sbjct 60 PLAKFNSVQ 68
> tpv:TP02_0735 translation initiation factor E4; K03259 translation
initiation factor 4E
Length=221
Score = 45.4 bits (106), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 36/67 (53%), Gaps = 3/67 (4%)
Query 78 LSFNPSVADAVNLDECITEER-PADAPMPLQHRWHVWEQIQREAAAADRAADYSQNTRDL 136
L+FN + D L E D P+ L+++W +WEQI + + DY ++T+ L
Sbjct 18 LTFNKNTEDHAKLSELFESTTINLDTPLSLKNKWVIWEQIVKMPEHSQN--DYKEHTKPL 75
Query 137 ASFDTVQ 143
SFD+VQ
Sbjct 76 VSFDSVQ 82
> bbo:BBOV_III011150 17.m10646; translation initiation factor
E4; K03259 translation initiation factor 4E
Length=242
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 35/67 (52%), Gaps = 2/67 (2%)
Query 78 LSFNPSVADAVNLDECI-TEERPADAPMPLQHRWHVWEQIQREAAAADRAADYSQNTRDL 136
L+FN D + E T PMPL+++W +WEQI + + + DY T+ L
Sbjct 22 LTFNKKSEDFKRVAELFETSNVDISTPMPLRNKWVIWEQIVK-GQDSRNSNDYKCYTKPL 80
Query 137 ASFDTVQ 143
SFD+VQ
Sbjct 81 VSFDSVQ 87
> mmu:244958 Mrap2, BB633055; melanocortin 2 receptor accessory
protein 2
Length=207
Score = 38.5 bits (88), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 27/48 (56%), Gaps = 0/48 (0%)
Query 3 FFVFFPLVLVLKRSGPEQEGARAADARFRRSSFASSAGGVCCSSPLFA 50
F+FF L L+ K P Q+ A +++ RFR +SF S G S +F+
Sbjct 60 IFMFFVLTLLTKTGAPHQDNAESSERRFRMNSFVSDFGKPLESDKVFS 107
> hsa:112609 MRAP2, C6orf117, RP11-51G5.2, bA51G5.2; melanocortin
2 receptor accessory protein 2
Length=205
Score = 37.4 bits (85), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 3 FFVFFPLVLVLKRSGPEQEGARAADARFRRSSFASSAG 40
F+FF L L+ K P Q+ A +++ RFR +SF S G
Sbjct 58 IFMFFVLTLLTKTGAPHQDNAESSEKRFRMNSFVSDFG 95
> dre:550549 eif4e1c, eif4E-1C, fb57e09, wu:fb57e09, zgc:110154;
eukaryotic translation initiation factor 4E family member
1c; K03259 translation initiation factor 4E
Length=213
Score = 32.7 bits (73), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 31/63 (49%), Gaps = 19/63 (30%)
Query 95 TEERPADAPM-------------PLQHRWHVWEQIQREAAAADRAADYSQNTRDLASFDT 141
TEE +D+P PLQ+RW +W D++ +++N R ++ FDT
Sbjct 11 TEEVRSDSPTAVVTTSPEQYIKHPLQNRWALW------YFKNDKSKSWTENLRLISKFDT 64
Query 142 VQD 144
V+D
Sbjct 65 VED 67
> dre:30738 eif4e1b, Z4ES, eif4e, zeIF4E; eukaryotic translation
initiation factor 4e 1b
Length=214
Score = 30.8 bits (68), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 21/40 (52%), Gaps = 6/40 (15%)
Query 105 PLQHRWHVWEQIQREAAAADRAADYSQNTRDLASFDTVQD 144
PLQ+RW +W D++ + N R + FDTV+D
Sbjct 35 PLQNRWGLW------FYKNDKSKMWQDNLRLITKFDTVED 68
> mmu:102098 Arhgef18, AI467246, D030053O22Rik; rho/rac guanine
nucleotide exchange factor (GEF) 18
Length=1021
Score = 30.0 bits (66), Expect = 4.7, Method: Composition-based stats.
Identities = 28/93 (30%), Positives = 37/93 (39%), Gaps = 16/93 (17%)
Query 91 DECITEERPADAPMPLQ---------HRWHVWEQIQREAAAADRAADY-------SQNTR 134
D E RPA + +P+Q + V +QI + AA+ + SQ
Sbjct 864 DSAPPESRPAKSDVPIQLLSATNQIQRQTAVQQQIPTKLAASTKGGKEKGSKSRGSQRWE 923
Query 135 DLASFDTVQDLGFRVFLLLRRSGSCGRTSPSPV 167
ASFD Q L F+ S S R S SPV
Sbjct 924 SSASFDLKQQLLLSKFIGKDESASRNRRSLSPV 956
> hsa:1977 EIF4E, CBP, EIF4E1, EIF4EL1, EIF4F, MGC111573; eukaryotic
translation initiation factor 4E; K03259 translation
initiation factor 4E
Length=237
Score = 29.6 bits (65), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 22/40 (55%), Gaps = 6/40 (15%)
Query 105 PLQHRWHVWEQIQREAAAADRAADYSQNTRDLASFDTVQD 144
PLQ+RW +W D++ + N R ++ FDTV+D
Sbjct 58 PLQNRWALW------FFKNDKSKTWQANLRLISKFDTVED 91
> xla:399255 eif4e, MGC84530; eIF-4E protein; K03259 translation
initiation factor 4E
Length=231
Score = 29.6 bits (65), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 22/40 (55%), Gaps = 6/40 (15%)
Query 105 PLQHRWHVWEQIQREAAAADRAADYSQNTRDLASFDTVQD 144
PLQ+RW +W D++ + N R ++ FDTV+D
Sbjct 52 PLQNRWALW------FFKNDKSKTWQANLRLISKFDTVED 85
> mmu:13684 Eif4e, EG668879, Eif4e-ps, If4e, MGC103177, eIF-4E;
eukaryotic translation initiation factor 4E; K03259 translation
initiation factor 4E
Length=217
Score = 29.6 bits (65), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 22/40 (55%), Gaps = 6/40 (15%)
Query 105 PLQHRWHVWEQIQREAAAADRAADYSQNTRDLASFDTVQD 144
PLQ+RW +W D++ + N R ++ FDTV+D
Sbjct 38 PLQNRWALW------FFKNDKSKTWQANLRLISKFDTVED 71
> xla:734259 eif4e, MGC85107, eIF-4E, eif4e1; eukaryotic translation
initiation factor 4E; K03259 translation initiation factor
4E
Length=213
Score = 29.6 bits (65), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 22/40 (55%), Gaps = 6/40 (15%)
Query 105 PLQHRWHVWEQIQREAAAADRAADYSQNTRDLASFDTVQD 144
PLQ+RW +W D++ + N R ++ FDTV+D
Sbjct 34 PLQNRWALW------FFKNDKSKTWQANLRLISKFDTVED 67
> dre:100332201 eukaryotic translation initiation factor 4E-1A-like
Length=215
Score = 29.6 bits (65), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 22/40 (55%), Gaps = 6/40 (15%)
Query 105 PLQHRWHVWEQIQREAAAADRAADYSQNTRDLASFDTVQD 144
PLQ+RW +W D++ + N R ++ FDTV+D
Sbjct 36 PLQNRWALW------FFKNDKSKTWQANLRLISKFDTVED 69
> tgo:TGME49_010780 ubiquitin carboxyl-terminal hydrolase, putative
(EC:3.1.2.15)
Length=4302
Score = 29.6 bits (65), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 29/76 (38%), Gaps = 16/76 (21%)
Query 66 CCCFWTMASTKYLSFNPSVADAVNLDECITEERPADAPMPLQHRWHV--WEQIQREAAAA 123
C CF ++ +LS I P D PM RWH WE + +A
Sbjct 3669 CACFLSLRMEDFLS--------------IAHGNPLDGPMAETPRWHPAEWEAVLGNMESA 3714
Query 124 DRAADYSQNTRDLASF 139
D+SQ+ + AS
Sbjct 3715 VLKIDFSQSLGNWASI 3730
> dre:79380 eif4e, eif4e-1, eif4e1a, zgc:86680; eukaryotic translation
initiation factor 4e; K03259 translation initiation
factor 4E
Length=215
Score = 29.3 bits (64), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 22/40 (55%), Gaps = 6/40 (15%)
Query 105 PLQHRWHVWEQIQREAAAADRAADYSQNTRDLASFDTVQD 144
PLQ+RW +W D++ + N R ++ FDTV+D
Sbjct 36 PLQNRWALW------FFKNDKSKTWQANLRLISKFDTVED 69
> dre:493617 zgc:101581
Length=216
Score = 29.3 bits (64), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 22/40 (55%), Gaps = 6/40 (15%)
Query 105 PLQHRWHVWEQIQREAAAADRAADYSQNTRDLASFDTVQD 144
PLQ+RW +W D++ + N R ++ FDTV+D
Sbjct 37 PLQNRWCLW------FFKNDKSKTWQANLRLISKFDTVED 70
Lambda K H
0.325 0.135 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4471152252
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40