bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3899_orf2
Length=111
Score E
Sequences producing significant alignments: (Bits) Value
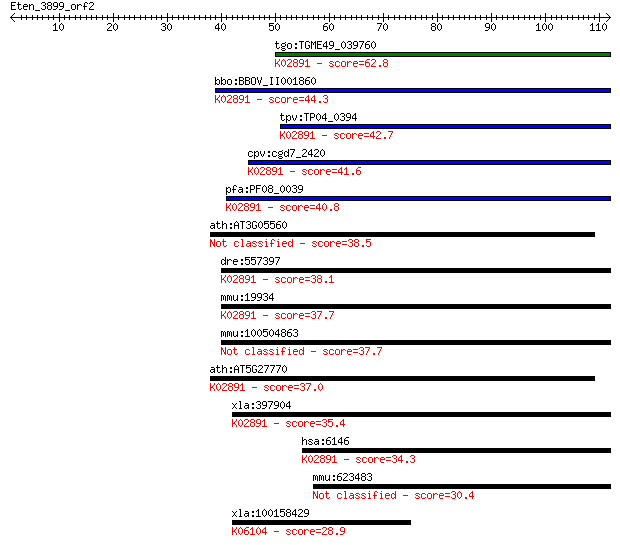
tgo:TGME49_039760 60S ribosomal protein L22, putative ; K02891... 62.8 3e-10
bbo:BBOV_II001860 18.m06142; 60S ribosomal L22e protein; K0289... 44.3 9e-05
tpv:TP04_0394 60S ribosomal protein L22; K02891 large subunit ... 42.7 3e-04
cpv:cgd7_2420 60S ribosomal protein L22 ; K02891 large subunit... 41.6 6e-04
pfa:PF08_0039 60S ribosomal protein L22, putative; K02891 larg... 40.8 0.001
ath:AT3G05560 60S ribosomal protein L22-2 (RPL22B) 38.5 0.005
dre:557397 MGC123327; zgc:123327; K02891 large subunit ribosom... 38.1 0.007
mmu:19934 Rpl22, 2700038K18Rik; ribosomal protein L22; K02891 ... 37.7 0.009
mmu:100504863 60S ribosomal protein L22-like 37.7 0.009
ath:AT5G27770 60S ribosomal protein L22 (RPL22C); K02891 large... 37.0 0.016
xla:397904 rpl22, xenla.hrpl22; ribosomal protein L22; K02891 ... 35.4 0.047
hsa:6146 RPL22, EAP, HBP15, HBP15/L22; ribosomal protein L22; ... 34.3 0.11
mmu:623483 Rpl22p; 60S ribosomal protein L22-like 30.4 1.6
xla:100158429 amotl1; angiomotin like 1; K06104 angiomotin like 1 28.9
> tgo:TGME49_039760 60S ribosomal protein L22, putative ; K02891
large subunit ribosomal protein L22e
Length=133
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 38/62 (61%), Gaps = 0/62 (0%)
Query 50 PPKIKFFLPPQNPVEENFFGAKGWGKFFQSPLKVGGGGTPFGEKIGVFRGRAKIWVPPGL 109
P K+KF + Q PV++N AKG +F Q+ +KV G G+++ V R +AK++V L
Sbjct 25 PVKVKFTVDCQKPVDDNIIEAKGLERFLQTHIKVDGKCNNLGDRVQVSREKAKVFVTAEL 84
Query 110 PF 111
PF
Sbjct 85 PF 86
> bbo:BBOV_II001860 18.m06142; 60S ribosomal L22e protein; K02891
large subunit ribosomal protein L22e
Length=122
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 32/73 (43%), Gaps = 2/73 (2%)
Query 39 SGGKKGPPPGKPPKIKFFLPPQNPVEENFFGAKGWGKFFQSPLKVGGGGTPFGEKIGVFR 98
+ K PG+ K KF L P +N G KF Q +KV G G I V R
Sbjct 7 ASNAKTIQPGQ--KSKFLLDCTAPANDNIINPSGLEKFLQDRIKVDGKTGNLGTNITVTR 64
Query 99 GRAKIWVPPGLPF 111
+ KI+V +PF
Sbjct 65 EKNKIYVVADIPF 77
> tpv:TP04_0394 60S ribosomal protein L22; K02891 large subunit
ribosomal protein L22e
Length=122
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 27/61 (44%), Gaps = 0/61 (0%)
Query 51 PKIKFFLPPQNPVEENFFGAKGWGKFFQSPLKVGGGGTPFGEKIGVFRGRAKIWVPPGLP 110
K+K+ L P +N G KF +KV G G K+ V R + KI V +P
Sbjct 17 HKVKYVLDCTGPANDNIINTAGLEKFLHDRIKVDGKTGNLGTKVLVTREKNKIHVTTEVP 76
Query 111 F 111
F
Sbjct 77 F 77
> cpv:cgd7_2420 60S ribosomal protein L22 ; K02891 large subunit
ribosomal protein L22e
Length=115
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 31/68 (45%), Gaps = 1/68 (1%)
Query 45 PPPGKPPKI-KFFLPPQNPVEENFFGAKGWGKFFQSPLKVGGGGTPFGEKIGVFRGRAKI 103
P K K +F + P+++N A G KFF +KV G G KI + R + +I
Sbjct 2 APITKAHKTQRFIVDCTAPMQDNIIDASGLEKFFHDRIKVDGKCGQLGTKIQISRQKGRI 61
Query 104 WVPPGLPF 111
V +P
Sbjct 62 TVLSEVPM 69
> pfa:PF08_0039 60S ribosomal protein L22, putative; K02891 large
subunit ribosomal protein L22e
Length=139
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/72 (27%), Positives = 30/72 (41%), Gaps = 1/72 (1%)
Query 41 GKKGPPPGKPPK-IKFFLPPQNPVEENFFGAKGWGKFFQSPLKVGGGGTPFGEKIGVFRG 99
G K K K IK+ L PV++ G +FF+ +KV K+ V
Sbjct 27 GLKKQKMNKSTKGIKYVLDCTKPVKDTILDISGLEQFFKDKIKVDKKTNNLKNKVVVTSD 86
Query 100 RAKIWVPPGLPF 111
KI++ +PF
Sbjct 87 EYKIYITVHIPF 98
> ath:AT3G05560 60S ribosomal protein L22-2 (RPL22B)
Length=124
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 30/71 (42%), Gaps = 0/71 (0%)
Query 38 ISGGKKGPPPGKPPKIKFFLPPQNPVEENFFGAKGWGKFFQSPLKVGGGGTPFGEKIGVF 97
+S G GK + F + PV++ KF Q +KVGG G+ + +
Sbjct 1 MSRGGAAVAKGKKKGVSFTIDCSKPVDDKIMEIASLEKFLQERIKVGGKAGALGDSVTIT 60
Query 98 RGRAKIWVPPG 108
R ++KI V
Sbjct 61 REKSKITVTAD 71
> dre:557397 MGC123327; zgc:123327; K02891 large subunit ribosomal
protein L22e
Length=127
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 36/73 (49%), Gaps = 6/73 (8%)
Query 40 GGKKGPPPGKPPKIKFFLPPQNPVEENFFGAKGWGKFFQSPLKVGG-GGTPFGEKIGVFR 98
GGKK K +KF L +PVE+ A + +F Q +KV G G G + + R
Sbjct 11 GGKK-----KKQVLKFTLDCTHPVEDGIMDAANFEQFLQERIKVNGKAGNLGGGVVSIER 65
Query 99 GRAKIWVPPGLPF 111
++KI V +PF
Sbjct 66 SKSKITVTSEVPF 78
> mmu:19934 Rpl22, 2700038K18Rik; ribosomal protein L22; K02891
large subunit ribosomal protein L22e
Length=128
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 36/73 (49%), Gaps = 6/73 (8%)
Query 40 GGKKGPPPGKPPKIKFFLPPQNPVEENFFGAKGWGKFFQSPLKVGG-GGTPFGEKIGVFR 98
GGKK K +KF L +PVE+ A + +F Q +KV G G G + + R
Sbjct 11 GGKK-----KKQVLKFTLDCTHPVEDGIMDAANFEQFLQERIKVNGKAGNLGGGVVTIER 65
Query 99 GRAKIWVPPGLPF 111
++KI V +PF
Sbjct 66 SKSKITVTSEVPF 78
> mmu:100504863 60S ribosomal protein L22-like
Length=128
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 36/73 (49%), Gaps = 6/73 (8%)
Query 40 GGKKGPPPGKPPKIKFFLPPQNPVEENFFGAKGWGKFFQSPLKVGG-GGTPFGEKIGVFR 98
GGKK K +KF L +PVE+ A + +F Q +KV G G G + + R
Sbjct 11 GGKK-----KKQVLKFTLDCTHPVEDGIMDAANFEQFLQERIKVNGKAGNLGGGVVTIER 65
Query 99 GRAKIWVPPGLPF 111
++KI V +PF
Sbjct 66 SKSKITVTSEVPF 78
> ath:AT5G27770 60S ribosomal protein L22 (RPL22C); K02891 large
subunit ribosomal protein L22e
Length=124
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 30/71 (42%), Gaps = 0/71 (0%)
Query 38 ISGGKKGPPPGKPPKIKFFLPPQNPVEENFFGAKGWGKFFQSPLKVGGGGTPFGEKIGVF 97
+S G GK + F + PV++ KF Q +KVGG G+ + +
Sbjct 1 MSRGNAAAAKGKKKGVSFTIDCSKPVDDKIMEIASLEKFLQERIKVGGKAGALGDSVSIT 60
Query 98 RGRAKIWVPPG 108
R ++KI V
Sbjct 61 REKSKITVTAD 71
> xla:397904 rpl22, xenla.hrpl22; ribosomal protein L22; K02891
large subunit ribosomal protein L22e
Length=128
Score = 35.4 bits (80), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 34/74 (45%), Gaps = 4/74 (5%)
Query 42 KKGPPPGKPPK---IKFFLPPQNPVEENFFGAKGWGKFFQSPLKVGGG-GTPFGEKIGVF 97
KK G K +KF L +PVE+ A + +F +KV G G G + +
Sbjct 5 KKTVTKGSKKKKQLLKFTLDCTHPVEDGIMDAANFEQFLHDRIKVNGKVGNLGGGVVSIE 64
Query 98 RGRAKIWVPPGLPF 111
R ++KI V +PF
Sbjct 65 RSKSKITVSSEVPF 78
> hsa:6146 RPL22, EAP, HBP15, HBP15/L22; ribosomal protein L22;
K02891 large subunit ribosomal protein L22e
Length=128
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 29/58 (50%), Gaps = 1/58 (1%)
Query 55 FFLPPQNPVEENFFGAKGWGKFFQSPLKVGG-GGTPFGEKIGVFRGRAKIWVPPGLPF 111
F L +PVE+ A + +F Q +KV G G G + + R ++KI V +PF
Sbjct 21 FTLDCTHPVEDGIMDAANFEQFLQERIKVNGKAGNLGGGVVTIERSKSKITVTSEVPF 78
> mmu:623483 Rpl22p; 60S ribosomal protein L22-like
Length=144
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 28/56 (50%), Gaps = 1/56 (1%)
Query 57 LPPQNPVEENFFGAKGWGKFFQSPLKVGG-GGTPFGEKIGVFRGRAKIWVPPGLPF 111
L +PVE+ A + +F Q +KV G G G + + + ++KI V +PF
Sbjct 27 LDCTHPVEDGIIDAANFEQFLQERIKVNGKAGNLGGGVVTIEQSKSKITVTSEVPF 82
> xla:100158429 amotl1; angiomotin like 1; K06104 angiomotin like
1
Length=862
Score = 28.9 bits (63), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 10/35 (28%), Positives = 20/35 (57%), Gaps = 2/35 (5%)
Query 42 KKGPPPGKPPKIKFFLPPQNPVEE--NFFGAKGWG 74
++GPPP P K+K + P N ++E +++ +
Sbjct 215 QRGPPPDYPSKVKHIVSPMNKMQEQGHYYNEQQHS 249
Lambda K H
0.317 0.148 0.494
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2062416360
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40