bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3967_orf1
Length=114
Score E
Sequences producing significant alignments: (Bits) Value
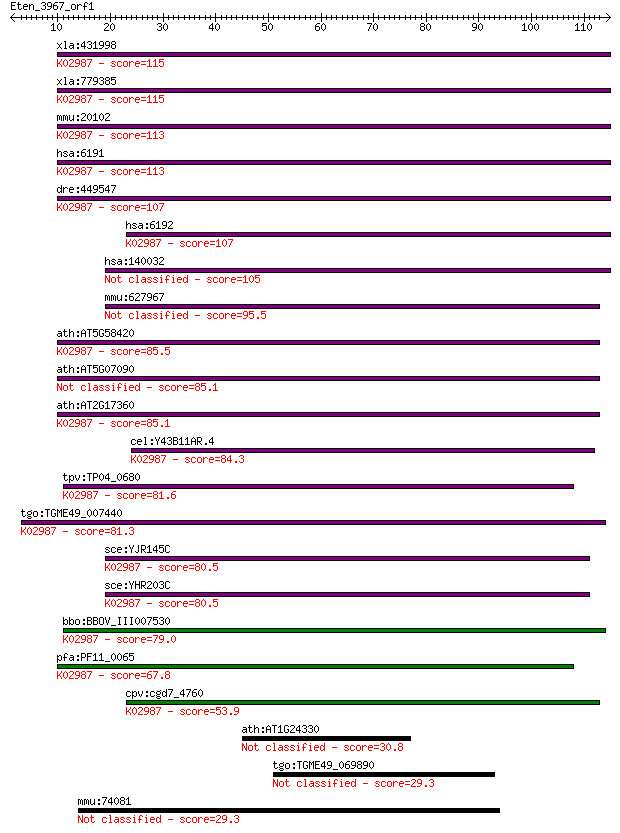
xla:431998 rps4, MGC81176; 40S ribosomal protein S4; K02987 sm... 115 3e-26
xla:779385 rps4x, MGC132392, ccg2, dxs306, rps4, scar, scr10; ... 115 3e-26
mmu:20102 Rps4x, MGC102174, MGC118104, Rps4, Rps4-1; ribosomal... 113 2e-25
hsa:6191 RPS4X, CCG2, DXS306, FLJ40595, RPS4, SCAR, SCR10; rib... 113 2e-25
dre:449547 rps4x, wu:fa91h09, zgc:92076; ribosomal protein S4,... 107 7e-24
hsa:6192 RPS4Y1, MGC119100, MGC5070, RPS4Y; ribosomal protein ... 107 1e-23
hsa:140032 RPS4Y2, RPS4Y2P; ribosomal protein S4, Y-linked 2 105 3e-23
mmu:627967 Gm6816, EG627967; predicted gene 6816 95.5
ath:AT5G58420 40S ribosomal protein S4 (RPS4D); K02987 small s... 85.5 4e-17
ath:AT5G07090 40S ribosomal protein S4 (RPS4B) 85.1 4e-17
ath:AT2G17360 40S ribosomal protein S4 (RPS4A); K02987 small s... 85.1 5e-17
cel:Y43B11AR.4 rps-4; Ribosomal Protein, Small subunit family ... 84.3 9e-17
tpv:TP04_0680 40S ribosomal protein S4; K02987 small subunit r... 81.6 6e-16
tgo:TGME49_007440 hypothetical protein ; K02987 small subunit ... 81.3 7e-16
sce:YJR145C RPS4A; Rps4ap; K02987 small subunit ribosomal prot... 80.5 1e-15
sce:YHR203C RPS4B; Rps4bp; K02987 small subunit ribosomal prot... 80.5 1e-15
bbo:BBOV_III007530 17.m07658; ribosomal protein S4e; K02987 sm... 79.0 3e-15
pfa:PF11_0065 40S ribosomal protein S4, putative; K02987 small... 67.8 8e-12
cpv:cgd7_4760 ribosomal protein S4 ; K02987 small subunit ribo... 53.9 1e-07
ath:AT1G24330 armadillo/beta-catenin repeat family protein / U... 30.8 1.1
tgo:TGME49_069890 M16 family peptidase, putative (EC:3.4.24.56) 29.3 3.3
mmu:74081 Cep350, 4933409L06Rik, 6430546F08Rik, MGC107324; cen... 29.3 3.6
> xla:431998 rps4, MGC81176; 40S ribosomal protein S4; K02987
small subunit ribosomal protein S4e
Length=263
Score = 115 bits (289), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 57/105 (54%), Positives = 68/105 (64%), Gaps = 0/105 (0%)
Query 10 PFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNF 69
++ + K + F KFD GN TGGAN G+IG I+N E PG FD+ H K ANGNNF
Sbjct 159 TIQIDLETGKITDFIKFDTGNLCMVTGGANLGRIGVITNRERHPGSFDVVHVKDANGNNF 218
Query 70 SPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQSNG 114
+ R SNIF GKGNKP + P GKGI L I E RDKRL +QS+G
Sbjct 219 ATRLSNIFVIGKGNKPWISLPRGKGIRLTIAEERDKRLAAKQSSG 263
> xla:779385 rps4x, MGC132392, ccg2, dxs306, rps4, scar, scr10;
ribosomal protein S4, X-linked; K02987 small subunit ribosomal
protein S4e
Length=263
Score = 115 bits (289), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 57/105 (54%), Positives = 68/105 (64%), Gaps = 0/105 (0%)
Query 10 PFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNF 69
++ + K + F KFD GN TGGAN G+IG I+N E PG FD+ H K ANGNNF
Sbjct 159 TIQIDLETGKITDFIKFDTGNLCMVTGGANLGRIGVITNRERHPGSFDVVHVKDANGNNF 218
Query 70 SPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQSNG 114
+ R SNIF GKGNKP + P GKGI L I E RDKRL +QS+G
Sbjct 219 ATRLSNIFVIGKGNKPWISLPRGKGIRLTIAEERDKRLAAKQSSG 263
> mmu:20102 Rps4x, MGC102174, MGC118104, Rps4, Rps4-1; ribosomal
protein S4, X-linked; K02987 small subunit ribosomal protein
S4e
Length=263
Score = 113 bits (282), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 56/105 (53%), Positives = 68/105 (64%), Gaps = 0/105 (0%)
Query 10 PFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNF 69
++ + K + F KFD GN TGGAN G+IG I+N E PG FD+ H K ANGN+F
Sbjct 159 TIQIDLETGKITDFIKFDTGNLCMVTGGANLGRIGVITNRERHPGSFDVVHVKDANGNSF 218
Query 70 SPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQSNG 114
+ R SNIF GKGNKP + P GKGI L I E RDKRL +QS+G
Sbjct 219 ATRLSNIFVIGKGNKPWISLPRGKGIRLTIAEERDKRLAAKQSSG 263
> hsa:6191 RPS4X, CCG2, DXS306, FLJ40595, RPS4, SCAR, SCR10; ribosomal
protein S4, X-linked; K02987 small subunit ribosomal
protein S4e
Length=263
Score = 113 bits (282), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 56/105 (53%), Positives = 68/105 (64%), Gaps = 0/105 (0%)
Query 10 PFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNF 69
++ + K + F KFD GN TGGAN G+IG I+N E PG FD+ H K ANGN+F
Sbjct 159 TIQIDLETGKITDFIKFDTGNLCMVTGGANLGRIGVITNRERHPGSFDVVHVKDANGNSF 218
Query 70 SPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQSNG 114
+ R SNIF GKGNKP + P GKGI L I E RDKRL +QS+G
Sbjct 219 ATRLSNIFVIGKGNKPWISLPRGKGIRLTIAEERDKRLAAKQSSG 263
> dre:449547 rps4x, wu:fa91h09, zgc:92076; ribosomal protein S4,
X-linked; K02987 small subunit ribosomal protein S4e
Length=263
Score = 107 bits (268), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 53/105 (50%), Positives = 65/105 (61%), Gaps = 0/105 (0%)
Query 10 PFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNF 69
R+ K + F KF+ GN TGGAN G+IG I+N E PG FD+ H K + GN+F
Sbjct 159 TIRIDLDTGKITDFIKFETGNMCMVTGGANLGRIGVITNREKHPGSFDVVHVKDSIGNSF 218
Query 70 SPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQSNG 114
+ R SNIF GKGNKP V P GKG+ L I E RDKRL +QS+
Sbjct 219 ATRLSNIFVIGKGNKPWVSLPRGKGVRLTIAEERDKRLAAKQSSS 263
> hsa:6192 RPS4Y1, MGC119100, MGC5070, RPS4Y; ribosomal protein
S4, Y-linked 1; K02987 small subunit ribosomal protein S4e
Length=263
Score = 107 bits (267), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 51/92 (55%), Positives = 61/92 (66%), Gaps = 0/92 (0%)
Query 23 FFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSPRFSNIFFFGKG 82
F KFD GN GGAN G++G I+N E PG FD+ H K ANGN+F+ R SNIF G G
Sbjct 172 FIKFDTGNLCMVIGGANLGRVGVITNRERHPGSFDVVHVKDANGNSFATRLSNIFVIGNG 231
Query 83 NKPGVFFPPGKGISLIIPEGRDKRLPPQQSNG 114
NKP + P GKGI L + E RDKRL +QS+G
Sbjct 232 NKPWISLPRGKGIRLTVAEERDKRLATKQSSG 263
> hsa:140032 RPS4Y2, RPS4Y2P; ribosomal protein S4, Y-linked 2
Length=263
Score = 105 bits (262), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 51/96 (53%), Positives = 61/96 (63%), Gaps = 0/96 (0%)
Query 19 KSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSPRFSNIFF 78
K + F KFD GN GAN G++G I+N E PG D+ H K ANGN+F+ R SNIF
Sbjct 168 KITSFIKFDTGNVCMVIAGANLGRVGVITNRERHPGSCDVVHVKDANGNSFATRISNIFV 227
Query 79 FGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQSNG 114
G GNKP + P GKGI L I E RDKRL +QS+G
Sbjct 228 IGNGNKPWISLPRGKGIRLTIAEERDKRLAAKQSSG 263
> mmu:627967 Gm6816, EG627967; predicted gene 6816
Length=255
Score = 95.5 bits (236), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 49/94 (52%), Positives = 56/94 (59%), Gaps = 0/94 (0%)
Query 19 KSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSPRFSNIFF 78
K + KFD GN TGGAN G+ I+N E P FD+ H K ANGN F+ R SNIF
Sbjct 161 KITDAIKFDTGNLGMVTGGANLGRTSVITNRERHPDSFDMVHMKDANGNGFATRLSNIFM 220
Query 79 FGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQS 112
KG KP + P GKGI L I E RDKRL +QS
Sbjct 221 IWKGKKPWISLPRGKGIRLTIAEERDKRLAAKQS 254
> ath:AT5G58420 40S ribosomal protein S4 (RPS4D); K02987 small
subunit ribosomal protein S4e
Length=262
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 43/103 (41%), Positives = 55/103 (53%), Gaps = 0/103 (0%)
Query 10 PFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNF 69
+L + K F KFD GN TGG N G++G I N E G F+ H + + G+ F
Sbjct 159 TIKLDLEANKIVEFIKFDVGNVVMVTGGRNRGRVGVIKNREKHKGSFETIHIQDSTGHEF 218
Query 70 SPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQS 112
+ R N++ GKG KP V P GKGI L I E KRL QQ+
Sbjct 219 ATRLGNVYTIGKGTKPWVSLPKGKGIKLTIIEEARKRLASQQA 261
> ath:AT5G07090 40S ribosomal protein S4 (RPS4B)
Length=244
Score = 85.1 bits (209), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 43/103 (41%), Positives = 55/103 (53%), Gaps = 0/103 (0%)
Query 10 PFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNF 69
+L + K F KFD GN TGG N G++G I N E G F+ H + + G+ F
Sbjct 141 TIKLDLEENKIVEFIKFDVGNVVMVTGGRNRGRVGVIKNREKHKGSFETIHIQDSTGHEF 200
Query 70 SPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQS 112
+ R N++ GKG KP V P GKGI L I E KRL QQ+
Sbjct 201 ATRLGNVYTIGKGTKPWVSLPKGKGIKLTIIEEARKRLASQQA 243
> ath:AT2G17360 40S ribosomal protein S4 (RPS4A); K02987 small
subunit ribosomal protein S4e
Length=261
Score = 85.1 bits (209), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 43/103 (41%), Positives = 55/103 (53%), Gaps = 0/103 (0%)
Query 10 PFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNF 69
+L + K F KFD GN TGG N G++G I N E G F+ H + + G+ F
Sbjct 159 TIKLDLEENKIVEFIKFDVGNVVMVTGGRNRGRVGVIKNREKHKGSFETIHIQDSTGHEF 218
Query 70 SPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQS 112
+ R N++ GKG KP V P GKGI L I E KRL QQ+
Sbjct 219 ATRLGNVYTIGKGTKPWVSLPKGKGIKLTIIEEARKRLSAQQA 261
> cel:Y43B11AR.4 rps-4; Ribosomal Protein, Small subunit family
member (rps-4); K02987 small subunit ribosomal protein S4e
Length=259
Score = 84.3 bits (207), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 41/88 (46%), Positives = 53/88 (60%), Gaps = 0/88 (0%)
Query 24 FKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSPRFSNIFFFGKGN 83
KF+PGN TGG N G++G I + E PG D+ H K + G++F+ R SN+F GKGN
Sbjct 172 VKFEPGNLAYVTGGRNVGRVGIIGHRERLPGASDIIHIKDSAGHSFATRISNVFVIGKGN 231
Query 84 KPGVFFPPGKGISLIIPEGRDKRLPPQQ 111
K V P G GI L I E RDKR+ +
Sbjct 232 KALVSLPTGAGIRLSIAEERDKRMAQKH 259
> tpv:TP04_0680 40S ribosomal protein S4; K02987 small subunit
ribosomal protein S4e
Length=262
Score = 81.6 bits (200), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 40/97 (41%), Positives = 52/97 (53%), Gaps = 0/97 (0%)
Query 11 FRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFS 70
R+ K F KF+PGN TGG N G++G + + E PG FDL H K + N FS
Sbjct 160 LRVEVSTGKVLEFLKFEPGNLVMITGGHNVGRVGTVVSKEKHPGSFDLVHVKDSQDNTFS 219
Query 71 PRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRL 107
R SN+F G G K V P +G+ I E R++RL
Sbjct 220 TRSSNVFVIGVGTKSYVSLPYERGLRKTIIEQRNERL 256
> tgo:TGME49_007440 hypothetical protein ; K02987 small subunit
ribosomal protein S4e
Length=301
Score = 81.3 bits (199), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 44/111 (39%), Positives = 54/111 (48%), Gaps = 0/111 (0%)
Query 3 PSSRGMIPFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWK 62
PS + RL K KF+ GN TGG N G++G I + E G FD+ H +
Sbjct 190 PSIKAHDCIRLDLNTGKIVDTLKFEAGNMAMVTGGHNVGRVGVIVHRERHLGGFDIIHLR 249
Query 63 KANGNNFSPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQSN 113
A N F+ R SN+F GKG K + P KGI L I E R L QQ N
Sbjct 250 DAKNNEFATRISNVFVIGKGEKAWISLPKEKGIRLSIMENRQVLLKKQQMN 300
> sce:YJR145C RPS4A; Rps4ap; K02987 small subunit ribosomal protein
S4e
Length=261
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 41/92 (44%), Positives = 50/92 (54%), Gaps = 0/92 (0%)
Query 19 KSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSPRFSNIFF 78
K + F KFD G TGG N G+IG I + E G FDL H K + N F R +N+F
Sbjct 168 KITDFIKFDAGKLVYVTGGRNLGRIGTIVHKERHDGGFDLVHIKDSLDNTFVTRLNNVFV 227
Query 79 FGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQ 110
G+ KP + P GKGI L I E RD+R Q
Sbjct 228 IGEQGKPYISLPKGKGIKLSIAEERDRRRAQQ 259
> sce:YHR203C RPS4B; Rps4bp; K02987 small subunit ribosomal protein
S4e
Length=261
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 41/92 (44%), Positives = 50/92 (54%), Gaps = 0/92 (0%)
Query 19 KSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSPRFSNIFF 78
K + F KFD G TGG N G+IG I + E G FDL H K + N F R +N+F
Sbjct 168 KITDFIKFDAGKLVYVTGGRNLGRIGTIVHKERHDGGFDLVHIKDSLDNTFVTRLNNVFV 227
Query 79 FGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQ 110
G+ KP + P GKGI L I E RD+R Q
Sbjct 228 IGEQGKPYISLPKGKGIKLSIAEERDRRRAQQ 259
> bbo:BBOV_III007530 17.m07658; ribosomal protein S4e; K02987
small subunit ribosomal protein S4e
Length=262
Score = 79.0 bits (193), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 42/103 (40%), Positives = 51/103 (49%), Gaps = 0/103 (0%)
Query 11 FRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFS 70
R+ + K FFKF+ GN TGG N G++G I + E PG FDL H + GNNF
Sbjct 160 LRIDLETGKVLEFFKFEVGNLVMITGGHNQGRVGTIVHKERHPGSFDLIHVRDELGNNFC 219
Query 71 PRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQSN 113
R SN+F G G V P +GI I E R RL N
Sbjct 220 TRCSNVFVIGVGTNNYVSLPKERGIKKGIIEDRADRLARASRN 262
> pfa:PF11_0065 40S ribosomal protein S4, putative; K02987 small
subunit ribosomal protein S4e
Length=261
Score = 67.8 bits (164), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 34/98 (34%), Positives = 47/98 (47%), Gaps = 0/98 (0%)
Query 10 PFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNF 69
RL + K KF G+ T G + G++G IS+ + G +D+ H K + F
Sbjct 159 TVRLDLETGKVLEHLKFQVGSLVMVTAGHSVGRVGVISSIDKNMGTYDIIHVKDSRNKVF 218
Query 70 SPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRL 107
+ R SN+F G KP + P KGI L I E R RL
Sbjct 219 ATRLSNVFVIGDNTKPYISLPREKGIKLDIIEERRNRL 256
> cpv:cgd7_4760 ribosomal protein S4 ; K02987 small subunit ribosomal
protein S4e
Length=260
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 43/90 (47%), Gaps = 0/90 (0%)
Query 23 FFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSPRFSNIFFFGKG 82
F K + GN TGG + G++G I++ E G + + G F+ NIF G+
Sbjct 171 FVKCEVGNMCMVTGGRSQGRVGTITHFERKMGAQCIVTIRDQKGATFATLMKNIFVIGEE 230
Query 83 NKPGVFFPPGKGISLIIPEGRDKRLPPQQS 112
KP V P KGI L E R+ R+ ++
Sbjct 231 GKPLVTLPKDKGIRLSNVEDRNLRMKKHRN 260
> ath:AT1G24330 armadillo/beta-catenin repeat family protein /
U-box domain-containing family protein
Length=771
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 12/32 (37%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 45 GISNPEGPPGFFDLAHWKKANGNNFSPRFSNI 76
GI+ P GPP DL +W+ A ++ SP ++
Sbjct 346 GITVPTGPPESLDLNYWRLAMSDSESPNSKSV 377
> tgo:TGME49_069890 M16 family peptidase, putative (EC:3.4.24.56)
Length=941
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 20/42 (47%), Gaps = 3/42 (7%)
Query 51 GPPGFFDLAHWKKANGNNFSPRFSNIFFFGKGNKPGVFFPPG 92
G FFD H N F+ +FS +F F G PG F PG
Sbjct 94 GTHDFFDFVHNHGGYTNAFTSKFSTVFSFSIG--PG-FLEPG 132
> mmu:74081 Cep350, 4933409L06Rik, 6430546F08Rik, MGC107324; centrosomal
protein 350
Length=3095
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 22/80 (27%), Positives = 32/80 (40%), Gaps = 2/80 (2%)
Query 14 PWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSPRF 73
P LA + F G+ G PG + + GF+ K GNN + +
Sbjct 2472 PLPLAATEELLDFHIGDRVL-IGSVQPGTLRFKGETDFAKGFWAGVELDKPEGNN-NGTY 2529
Query 74 SNIFFFGKGNKPGVFFPPGK 93
I +F +K G+F PP K
Sbjct 2530 DGIVYFVCKDKHGIFAPPQK 2549
Lambda K H
0.320 0.146 0.483
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2047611720
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40