bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4008_orf1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
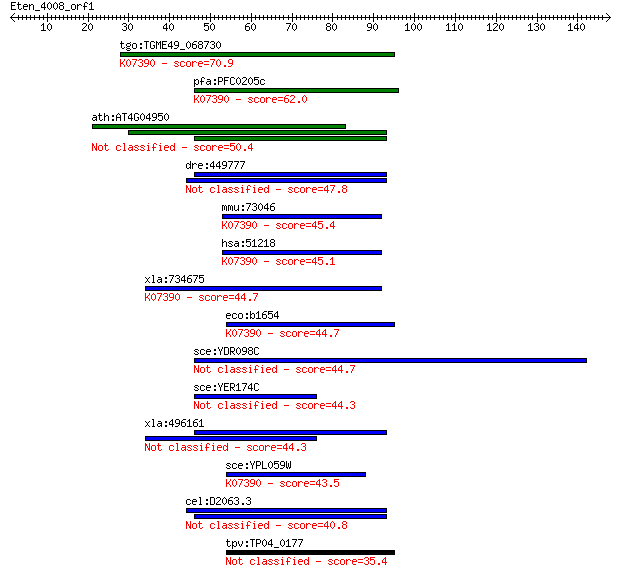
tgo:TGME49_068730 hypothetical protein ; K07390 monothiol glut... 70.9 1e-12
pfa:PFC0205c PfGLP-1; 1-cys-glutaredoxin-like protein-1; K0739... 62.0 7e-10
ath:AT4G04950 thioredoxin family protein 50.4 2e-06
dre:449777 glrx3, txnl2, wu:fb38e09, zgc:103648; glutaredoxin 3 47.8
mmu:73046 Glrx5, 2310004O13Rik, 2900070E19Rik, AU020725; gluta... 45.4 6e-05
hsa:51218 GLRX5, C14orf87, FLB4739, GRX5, MGC14129, PR01238, P... 45.1 9e-05
xla:734675 glrx5, MGC116473, grx5; glutaredoxin 5; K07390 mono... 44.7 9e-05
eco:b1654 grxD, ECK1650, JW1646, ydhD; glutaredoxin-4; K07390 ... 44.7 1e-04
sce:YDR098C GRX3; Grx3p 44.7 1e-04
sce:YER174C GRX4; Grx4p 44.3 1e-04
xla:496161 glrx3, txnl2; glutaredoxin 3 44.3
sce:YPL059W GRX5; Grx5p; K07390 monothiol glutaredoxin 43.5 3e-04
cel:D2063.3 hypothetical protein 40.8 0.002
tpv:TP04_0177 hypothetical protein 35.4 0.061
> tgo:TGME49_068730 hypothetical protein ; K07390 monothiol glutaredoxin
Length=138
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 34/67 (50%), Positives = 46/67 (68%), Gaps = 0/67 (0%)
Query 28 RGTQQNSKTSASPMSDEELEAKLLQLVEKTPVVLFMKGRPEAPLCGFSARALLLLEAAGV 87
R T+ + P ++L+ KL LV+ TPVVLFMKG P+AP+CGFSARA+ +L + V
Sbjct 2 RRTRCQFRAYVWPDFFQDLKKKLDDLVKSTPVVLFMKGSPKAPMCGFSARAVGILNSLDV 61
Query 88 REYTFVD 94
EYTFV+
Sbjct 62 DEYTFVN 68
> pfa:PFC0205c PfGLP-1; 1-cys-glutaredoxin-like protein-1; K07390
monothiol glutaredoxin
Length=171
Score = 62.0 bits (149), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 23/50 (46%), Positives = 36/50 (72%), Gaps = 0/50 (0%)
Query 46 LEAKLLQLVEKTPVVLFMKGRPEAPLCGFSARALLLLEAAGVREYTFVDC 95
L+ K+ +L+E+ +VLFMKG PE PLCGFSA + +L + V++Y ++D
Sbjct 73 LKIKIKELLEQEKIVLFMKGTPEKPLCGFSANVVNILNSMNVKDYVYIDV 122
> ath:AT4G04950 thioredoxin family protein
Length=488
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 26/62 (41%), Positives = 37/62 (59%), Gaps = 1/62 (1%)
Query 21 QQQQQEERGTQQNSKTSASPMSDEELEAKLLQLVEKTPVVLFMKGRPEAPLCGFSARALL 80
++ Q+E G + + +S E L A+L LV PV+LFMKGRPE P CGFS + +
Sbjct 259 SKESQDEAGKGGGVSSGNTGLS-ETLRARLEGLVNSKPVMLFMKGRPEEPKCGFSGKVVE 317
Query 81 LL 82
+L
Sbjct 318 IL 319
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 26/68 (38%), Positives = 41/68 (60%), Gaps = 5/68 (7%)
Query 30 TQQNSKTS----ASPMSDEE-LEAKLLQLVEKTPVVLFMKGRPEAPLCGFSARALLLLEA 84
++N+K S A P+S + L+++L +L PV+LFMKG PE P CGFS + + +L+
Sbjct 132 VKENAKASLQDRAQPVSTADALKSRLEKLTNSHPVMLFMKGIPEEPRCGFSRKVVDILKE 191
Query 85 AGVREYTF 92
V +F
Sbjct 192 VNVDFGSF 199
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 28/47 (59%), Gaps = 0/47 (0%)
Query 46 LEAKLLQLVEKTPVVLFMKGRPEAPLCGFSARALLLLEAAGVREYTF 92
LE +L L+ + V+LFMKG P+ P CGFS++ + L V +F
Sbjct 390 LEDRLKALINSSEVMLFMKGSPDEPKCGFSSKVVKALRGENVSFGSF 436
> dre:449777 glrx3, txnl2, wu:fb38e09, zgc:103648; glutaredoxin
3
Length=326
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/47 (48%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 46 LEAKLLQLVEKTPVVLFMKGRPEAPLCGFSARALLLLEAAGVREYTF 92
LE +L L+ K+PV+LFMKG EA CGFS + L ++ GV TF
Sbjct 226 LENRLKSLINKSPVMLFMKGNKEAAKCGFSRQILEIMNNTGVEYDTF 272
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 31/49 (63%), Gaps = 0/49 (0%)
Query 44 EELEAKLLQLVEKTPVVLFMKGRPEAPLCGFSARALLLLEAAGVREYTF 92
E+L +L +L+ P +LFMKG P+ P CGFS + + +L+ V+ +F
Sbjct 122 EDLNQRLKRLINAAPCMLFMKGSPQEPRCGFSRQIIQILKDHNVQYSSF 170
> mmu:73046 Glrx5, 2310004O13Rik, 2900070E19Rik, AU020725; glutaredoxin
5 homolog (S. cerevisiae); K07390 monothiol glutaredoxin
Length=152
Score = 45.4 bits (106), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 26/39 (66%), Gaps = 0/39 (0%)
Query 53 LVEKTPVVLFMKGRPEAPLCGFSARALLLLEAAGVREYT 91
LV+K VV+F+KG PE P CGFS + +L GVR+Y
Sbjct 44 LVKKDKVVVFLKGTPEQPQCGFSNAVVQILRLHGVRDYA 82
> hsa:51218 GLRX5, C14orf87, FLB4739, GRX5, MGC14129, PR01238,
PRO1238; glutaredoxin 5; K07390 monothiol glutaredoxin
Length=157
Score = 45.1 bits (105), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 26/39 (66%), Gaps = 0/39 (0%)
Query 53 LVEKTPVVLFMKGRPEAPLCGFSARALLLLEAAGVREYT 91
LV+K VV+F+KG PE P CGFS + +L GVR+Y
Sbjct 48 LVKKDKVVVFLKGTPEQPQCGFSNAVVQILRLHGVRDYA 86
> xla:734675 glrx5, MGC116473, grx5; glutaredoxin 5; K07390 monothiol
glutaredoxin
Length=154
Score = 44.7 bits (104), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 24/58 (41%), Positives = 32/58 (55%), Gaps = 7/58 (12%)
Query 34 SKTSASPMSDEELEAKLLQLVEKTPVVLFMKGRPEAPLCGFSARALLLLEAAGVREYT 91
S S SP E L+ LV+K VV+F+KG P P+CGFS + +L GV +Y
Sbjct 33 SSDSVSP---EHLDG----LVKKDKVVVFIKGTPAQPMCGFSNAVVQILRMHGVNDYA 83
> eco:b1654 grxD, ECK1650, JW1646, ydhD; glutaredoxin-4; K07390
monothiol glutaredoxin
Length=115
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 29/41 (70%), Gaps = 1/41 (2%)
Query 54 VEKTPVVLFMKGRPEAPLCGFSARALLLLEAAGVREYTFVD 94
+ + P++L+MKG P+ P CGFSA+A+ L A G R + +VD
Sbjct 12 IAENPILLYMKGSPKLPSCGFSAQAVQALAACGER-FAYVD 51
> sce:YDR098C GRX3; Grx3p
Length=285
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 34/102 (33%), Positives = 50/102 (49%), Gaps = 7/102 (6%)
Query 46 LEAKLLQLVEKTPVVLFMKGRPEAPLCGFSARALLLLEAAGVREYTFVDC----SIPQHL 101
+ A+L +LV PV+LFMKG P P CGFS + + +L VR + F D S+ Q+L
Sbjct 185 INARLTKLVNAAPVMLFMKGSPSEPKCGFSRQLVGILREHQVR-FGFFDILRDESVRQNL 243
Query 102 PQQQQQQQ--QQQRQGQQRQGQERKKQQGGEGEDSEQQQLQQ 141
+ + Q G+ + G + K+ E D Q LQ
Sbjct 244 KKFSEWPTFPQLYINGEFQGGLDIIKESLEEDPDFLQHALQS 285
> sce:YER174C GRX4; Grx4p
Length=244
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 17/30 (56%), Positives = 23/30 (76%), Gaps = 0/30 (0%)
Query 46 LEAKLLQLVEKTPVVLFMKGRPEAPLCGFS 75
+ A+L++LV+ PV+LFMKG P P CGFS
Sbjct 145 INARLVKLVQAAPVMLFMKGSPSEPKCGFS 174
> xla:496161 glrx3, txnl2; glutaredoxin 3
Length=326
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 23/47 (48%), Positives = 28/47 (59%), Gaps = 0/47 (0%)
Query 46 LEAKLLQLVEKTPVVLFMKGRPEAPLCGFSARALLLLEAAGVREYTF 92
LE +L LV K PV+LFMKG E CGFS + L ++ GV TF
Sbjct 226 LEERLKVLVNKAPVMLFMKGNKEMAKCGFSRQILEIMNNTGVNFETF 272
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 21/46 (45%), Positives = 29/46 (63%), Gaps = 4/46 (8%)
Query 34 SKTS--ASPMS--DEELEAKLLQLVEKTPVVLFMKGRPEAPLCGFS 75
S TS A+P S E+L +L +L+ P +LF+KG P+ P CGFS
Sbjct 108 SSTSFPATPNSAPKEDLNGRLKKLINAAPCMLFIKGSPQEPRCGFS 153
> sce:YPL059W GRX5; Grx5p; K07390 monothiol glutaredoxin
Length=150
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 20/34 (58%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 54 VEKTPVVLFMKGRPEAPLCGFSARALLLLEAAGV 87
+E PVVLFMKG PE P CGFS + LL GV
Sbjct 42 IESAPVVLFMKGTPEFPKCGFSRATIGLLGNQGV 75
> cel:D2063.3 hypothetical protein
Length=342
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 21/49 (42%), Positives = 29/49 (59%), Gaps = 0/49 (0%)
Query 44 EELEAKLLQLVEKTPVVLFMKGRPEAPLCGFSARALLLLEAAGVREYTF 92
E L A+L LV V++FMKG P AP CGFS + LL + ++ +F
Sbjct 133 EALNARLGALVNSQKVMVFMKGDPSAPRCGFSRTIVELLNSHKIKFGSF 181
Score = 35.8 bits (81), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 21/47 (44%), Positives = 27/47 (57%), Gaps = 0/47 (0%)
Query 46 LEAKLLQLVEKTPVVLFMKGRPEAPLCGFSARALLLLEAAGVREYTF 92
LE +L +LV ++LFMKG E P CGFS + LL A +TF
Sbjct 243 LEDRLKKLVSSQRLMLFMKGDRETPKCGFSRTIVDLLNKARADYHTF 289
> tpv:TP04_0177 hypothetical protein
Length=159
Score = 35.4 bits (80), Expect = 0.061, Method: Compositional matrix adjust.
Identities = 12/41 (29%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 54 VEKTPVVLFMKGRPEAPLCGFSARALLLLEAAGVREYTFVD 94
+E +++F+KG PE P C +SA+ + +L + +Y +++
Sbjct 66 IETEKILIFIKGTPEDPQCKYSAKLIEILNNYKLNDYAYIN 106
Lambda K H
0.307 0.121 0.320
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40