bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4089_orf1
Length=240
Score E
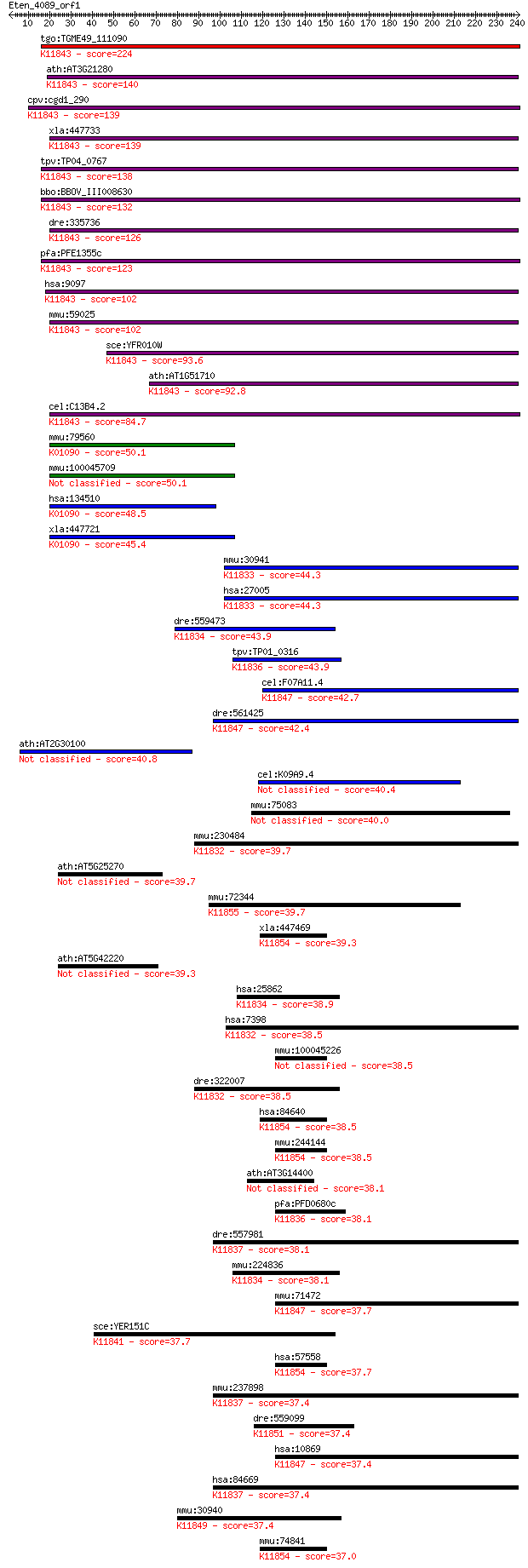
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_111090 ubiquitin carboxyl-terminal hydrolase, putat... 224 2e-58
ath:AT3G21280 UBP7; UBP7 (UBIQUITIN-SPECIFIC PROTEASE 7); ubiq... 140 3e-33
cpv:cgd1_290 Ub6p like ubiquitin at N-terminus and ubiquitin C... 139 6e-33
xla:447733 usp14, MGC81945; ubiquitin specific peptidase 14 (t... 139 1e-32
tpv:TP04_0767 ubiquitin carboxyl-terminal hydrolase (EC:3.1.2.... 138 1e-32
bbo:BBOV_III008630 17.m07754; ubiquitin carboxyl-terminal hydr... 132 1e-30
dre:335736 usp14, wu:fk63d09, zgc:55949; ubiquitin specific pr... 126 5e-29
pfa:PFE1355c ubiquitin carboxyl-terminal hydrolase, putative (... 123 5e-28
hsa:9097 USP14, TGT; ubiquitin specific peptidase 14 (tRNA-gua... 102 1e-21
mmu:59025 Usp14, 2610005K12Rik, 2610037B11Rik, AW107924, C7876... 102 1e-21
sce:YFR010W UBP6; Ubiquitin-specific protease situated in the ... 93.6 6e-19
ath:AT1G51710 UBP6; UBP6 (UBIQUITIN-SPECIFIC PROTEASE 6); calm... 92.8 9e-19
cel:C13B4.2 usp-14; Ubiquitin Specific Protease family member ... 84.7 3e-16
mmu:79560 Ublcp1, 4930527B16Rik, 8430435I17Rik, BC002236, MGC1... 50.1 7e-06
mmu:100045709 ubiquitin-like domain-containing CTD phosphatase... 50.1 8e-06
hsa:134510 UBLCP1, CPUB1, FLJ25267, MGC10067; ubiquitin-like d... 48.5 2e-05
xla:447721 ublcp1; ubiquitin-like domain containing CTD phosph... 45.4 2e-04
mmu:30941 Usp21, ESTM28, Usp16, Usp23, W53272; ubiquitin speci... 44.3 4e-04
hsa:27005 USP21, MGC3394, USP16, USP23; ubiquitin specific pep... 44.3 4e-04
dre:559473 usp49, si:rp71-11b17.7; ubiquitin specific peptidas... 43.9 5e-04
tpv:TP01_0316 ubiquitin carboxyl-terminal hydrolase a; K11836 ... 43.9 5e-04
cel:F07A11.4 hypothetical protein; K11847 ubiquitin carboxyl-t... 42.7 0.001
dre:561425 novel protein similar to vertebrate ubiquitin speci... 42.4 0.001
ath:AT2G30100 ubiquitin family protein 40.8 0.004
cel:K09A9.4 hypothetical protein 40.4 0.005
mmu:75083 Usp50, 1700086G18Rik, 4930511O11Rik; ubiquitin speci... 40.0 0.007
mmu:230484 Usp1, MGC25559; ubiquitin specific peptidase 1 (EC:... 39.7 0.009
ath:AT5G25270 hypothetical protein 39.7 0.011
mmu:72344 Usp36, 2700002L06Rik, mKIAA1453; ubiquitin specific ... 39.7 0.011
xla:447469 usp38, MGC81730; ubiquitin specific peptidase 38 (E... 39.3 0.013
ath:AT5G42220 ubiquitin family protein 39.3 0.014
hsa:25862 USP49, MGC20741; ubiquitin specific peptidase 49 (EC... 38.9 0.015
hsa:7398 USP1, UBP; ubiquitin specific peptidase 1 (EC:3.4.19.... 38.5 0.020
mmu:100045226 ubiquitin carboxyl-terminal hydrolase 35-like 38.5 0.022
dre:322007 usp1, wu:fb43a02, zgc:55962; ubiquitin specific pro... 38.5 0.023
hsa:84640 USP38, FLJ35970, HP43.8KD, KIAA1891; ubiquitin speci... 38.5 0.024
mmu:244144 Usp35, Gm1088, Gm493; ubiquitin specific peptidase ... 38.5 0.024
ath:AT3G14400 UBP25; UBP25 (UBIQUITIN-SPECIFIC PROTEASE 25); u... 38.1 0.025
pfa:PFD0680c ubiquitin carboxyl-terminal hydrolase A, putative... 38.1 0.027
dre:557981 ubiquitin specific peptidase 15-like; K11837 ubiqui... 38.1 0.029
mmu:224836 Usp49, C330046L10Rik, Gm545; ubiquitin specific pep... 38.1 0.029
mmu:71472 Usp19, 8430421I07Rik, AI047774, Zmynd9; ubiquitin sp... 37.7 0.033
sce:YER151C UBP3, BLM3; Ubiquitin-specific protease that inter... 37.7 0.034
hsa:57558 USP35; ubiquitin specific peptidase 35 (EC:3.4.19.12... 37.7 0.034
mmu:237898 Usp32, 2900074J03Rik, 6430526O11Rik, AW045245; ubiq... 37.4 0.044
dre:559099 usp30, si:dkey-72n1.2; ubiquitin specific peptidase... 37.4 0.047
hsa:10869 USP19, ZMYND9; ubiquitin specific peptidase 19 (EC:3... 37.4 0.048
hsa:84669 USP32, NY-REN-60, USP10; ubiquitin specific peptidas... 37.4 0.054
mmu:30940 Usp25; ubiquitin specific peptidase 25 (EC:3.4.19.12... 37.4 0.055
mmu:74841 Usp38, 4631402N15Rik, 4833420O05Rik, AA536967, AU044... 37.0 0.056
> tgo:TGME49_111090 ubiquitin carboxyl-terminal hydrolase, putative
(EC:3.1.2.15 3.1.3.16); K11843 ubiquitin carboxyl-terminal
hydrolase 14 [EC:3.1.2.15]
Length=607
Score = 224 bits (571), Expect = 2e-58, Method: Compositional matrix adjust.
Identities = 120/231 (51%), Positives = 160/231 (69%), Gaps = 7/231 (3%)
Query 16 VQAAVKWGAKSF-NVQIDPNEPLEVFQAQLYTLTGVPVERQKLLCKGKTIKVDADLQALV 74
++ +VKW + F NV++D +EPL + AQL TLTGVP +RQKL+ K +K DL+A
Sbjct 4 IRISVKWNKQEFENVEVDLSEPLALLNAQLMTLTGVPADRQKLMANRKLVKTVDDLRAAA 63
Query 75 SSGCP-KIMLMGTAEEKTAVKPPPEKTVFVEDLTPAQQAALLKEKKVEPLPNGIVNLGNT 133
G ++ ++GTAE ++ P E+TVFVEDLTPA++A LL+EK +EPLP GI NLGNT
Sbjct 64 KGGKNLRVTMLGTAE-GAELQQPLERTVFVEDLTPAERAKLLREKNIEPLPPGITNLGNT 122
Query 134 CYLASVLQMLRPAKDFSDLLKNYI---SGGAATAQFAATGQQGALRRLSLALKDFYNQWD 190
CYLASVLQMLRP ++ + +KN + +G +A A A +G GA RL+LALKDF++ WD
Sbjct 123 CYLASVLQMLRPCRELTAAVKNDMKNSTGESAAAYTALSGAVGAAYRLALALKDFHSHWD 182
Query 191 QTVEAVSPFLLVPTLREAYPQFSRRSTSQAGTTGVP-MQQDAEECLSCLMT 240
T A P LLV LREA+PQF+R++ + G G MQQDAEECLSCL+
Sbjct 183 STAGAAQPVLLVHALREAFPQFARKAAAGPGGLGGGFMQQDAEECLSCLVN 233
> ath:AT3G21280 UBP7; UBP7 (UBIQUITIN-SPECIFIC PROTEASE 7); ubiquitin
thiolesterase/ ubiquitin-specific protease; K11843 ubiquitin
carboxyl-terminal hydrolase 14 [EC:3.1.2.15]
Length=532
Score = 140 bits (354), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 89/222 (40%), Positives = 127/222 (57%), Gaps = 22/222 (9%)
Query 19 AVKWGAKSF-NVQIDPNEPLEVFQAQLYTLTGVPVERQKLLCKGKTIKVDADLQALVSSG 77
+VKW K F +++ID ++P VF+AQLY L+GVP ERQK++ KG +K DAD L
Sbjct 60 SVKWQKKVFESIEIDTSQPPFVFKAQLYDLSGVPPERQKIMVKGGLLKDDADWSTLGLKN 119
Query 78 CPKIMLMGTAEEKTAVKPPPEKTVFVEDLTPAQQAALLKEKKVEPLPNGIVNLGNTCYLA 137
K+M+MGTA+E VK P + VF+EDL QQAA L G+VNLGNTCY+
Sbjct 120 GQKLMMMGTADE--IVKAPEKGPVFMEDLPEEQQAANLG------YSAGLVNLGNTCYMN 171
Query 138 SVLQMLRPAKDFSDLLKNYISGGAATAQFAATGQQGALRRLSLALKDFYNQWDQTVEAVS 197
S +Q L + L NY S A T T L++A ++ +++ D++V+AV+
Sbjct 172 STMQCLISVPELKSELSNYQS--ARTKDVDQTSHM-----LTVATRELFSELDKSVKAVA 224
Query 198 PFLLVPTLREAYPQFSRRSTSQAGTTGVPMQQDAEECLSCLM 239
P L++ YPQF++ G MQQDAEEC + ++
Sbjct 225 PMPFWMVLQKKYPQFAQLH------NGNHMQQDAEECWTQML 260
> cpv:cgd1_290 Ub6p like ubiquitin at N-terminus and ubiquitin
C terminal hydrolase at the C-terminus ; K11843 ubiquitin carboxyl-terminal
hydrolase 14 [EC:3.1.2.15]
Length=499
Score = 139 bits (351), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 86/233 (36%), Positives = 134/233 (57%), Gaps = 10/233 (4%)
Query 10 KMKASE-VQAAVKWGAKSF-NVQIDPNEPLEVFQAQLYTLTGVPVERQKLLCKGKTIKVD 67
K+ A E ++ VKW K+F +V + + PL F+ +L LTGVP +QKL+ + ++
Sbjct 8 KLNAMEQIKVTVKWNVKTFEDVDLVFSSPLSEFRERLSNLTGVPASKQKLMSQRGVLRDG 67
Query 68 ADLQALVSSGCPKIMLMGTAEEKTAVKPPPEKTVFVEDLTPAQQAALLKEKKVEPLPNGI 127
DL L KI+L+GTAE +K P EKT+F EDLT ++A +L EK++ PLP G+
Sbjct 68 MDLNKLGLKPGSKIVLVGTAE-GGELKAPSEKTIFFEDLTSDERAKILHEKEIVPLPVGL 126
Query 128 VNLGNTCYLASVLQMLRPAKDFSDLLKNYISGGAATAQFAATGQQGALRRLSLALKDFYN 187
NLGNTCYL S++ MLR F +L+N ++++ ++ +++ L+ + K +
Sbjct 127 ENLGNTCYLNSIIHMLRSIPSFLQILRN---SNLSSSERTSSSSGNSIKFLN-SFKQLMD 182
Query 188 QWDQTVEAVSPFLLVPTLREAYPQFSRRSTSQAGTTGVPMQQDAEECLSCLMT 240
+ D ++E V P V R +PQ+ ST+ G + QQDAEE L L+T
Sbjct 183 KMDGSIERVVPGECVDLFRRQFPQY---STTTGGAFAMYQQQDAEEVLGSLLT 232
> xla:447733 usp14, MGC81945; ubiquitin specific peptidase 14
(tRNA-guanine transglycosylase) (EC:3.1.2.15); K11843 ubiquitin
carboxyl-terminal hydrolase 14 [EC:3.1.2.15]
Length=489
Score = 139 bits (349), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 83/224 (37%), Positives = 131/224 (58%), Gaps = 27/224 (12%)
Query 20 VKWGAKSF-NVQIDPNEPLEVFQAQLYTLTGVPVERQKLLCKGKTIKVDADLQALVSSGC 78
VKWG + F NV+++ +EP VF+AQL+ LTGV +RQK++ KG T+K D + SG
Sbjct 8 VKWGKEKFDNVELNTDEPPMVFKAQLFALTGVQPDRQKVMVKGGTLKDDDWGNLKIKSGM 67
Query 79 PKIMLMGTAE---EKTAVKPPPEKTVFVEDLTPAQQAALLKEKKVEPLPNGIVNLGNTCY 135
+++MG+AE E+ AV+P VFVED+T Q A+ ++ LP G+ NLGNTCY
Sbjct 68 -TLLMMGSAEALPEEPAVRP-----VFVEDMTEEQLASAME------LPCGLTNLGNTCY 115
Query 136 LASVLQMLRPAKDFSDLLKNYISGGAATAQFAATGQQGALRRLSLALKDFYNQWDQTVEA 195
+ + +Q +R + + LK Y T A+G+ + + ++ AL+D ++ D+T +
Sbjct 116 MNATVQCIRSVPELKEALKRY------TGALRASGEMASAQYITAALRDLFDSMDKTSSS 169
Query 196 VSPFLLVPTLREAYPQFSRRSTSQAGTTGVPMQQDAEECLSCLM 239
+ P +L+ L A+PQF+ + G G +QQDA EC +M
Sbjct 170 IPPIILLQFLHMAFPQFAEK-----GDQGQYLQQDANECWVQVM 208
> tpv:TP04_0767 ubiquitin carboxyl-terminal hydrolase (EC:3.1.2.15);
K11843 ubiquitin carboxyl-terminal hydrolase 14 [EC:3.1.2.15]
Length=526
Score = 138 bits (348), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 85/229 (37%), Positives = 127/229 (55%), Gaps = 27/229 (11%)
Query 16 VQAAVKWGAKSF-NVQIDPNEPLEVFQAQLYTLTGVPVERQKLLCKGKTIKVDADLQALV 74
V VKW K + +++ +EPLE F+ QL +LTGVP ERQK++ KG I DADL +
Sbjct 8 VSVNVKWMGKQYEGLKMSLDEPLESFKNQLCSLTGVPPERQKIMFKG-IIPNDADLSKIK 66
Query 75 SSGCPKIMLMGTAEEKTAVKPPP--EKTVFVEDLTPAQQAALLKEKKVEPLPNGIVNLGN 132
+ ++ML+G+AE KPP EK F E+L+ ++A ++ + + LP GI+NLGN
Sbjct 67 ITNGARLMLIGSAE-----KPPECIEKVRFFEELSSQEKAKIMNDNIIVKLPPGIMNLGN 121
Query 133 TCYLASVLQMLRPAKDFSDLLKN--YISGGAATAQFAATGQQGALRRLSLALKDFYNQWD 190
TCY SV+Q L P + + + ++G + FA +L D NQ +
Sbjct 122 TCYFNSVVQFLFPVTELWNSVSKCLEVNGNSPDVHFAK------------SLLDMKNQLN 169
Query 191 QTVEAVSPFLLVPTLREAYPQFSRRSTSQAGTTGVPMQQDAEECLSCLM 239
T+E P + + LR+ P F R+ TG+ MQQDAEECL+C++
Sbjct 170 HTLERFVPLVQIQFLRKINPLFCRKDEK----TGMYMQQDAEECLNCIL 214
> bbo:BBOV_III008630 17.m07754; ubiquitin carboxyl-terminal hydrolase
family protein; K11843 ubiquitin carboxyl-terminal hydrolase
14 [EC:3.1.2.15]
Length=496
Score = 132 bits (332), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 84/229 (36%), Positives = 124/229 (54%), Gaps = 22/229 (9%)
Query 16 VQAAVKWGAKSFN-VQIDPNEPLEVFQAQLYTLTGVPVERQKLLCKGKTIKVDADLQALV 74
V+ VKW K FN ++++ +E LE+F+ QL++LTGVP ERQKL+ KG + DL+
Sbjct 8 VRVTVKWMGKQFNDLELNLSESLELFRVQLFSLTGVPPERQKLMFKG-LLSDSIDLRNTG 66
Query 75 SSGCPKIMLMGTAEEKTAVKPPPEKTVFVEDLTPAQQAALLKEKKVEPLPNGIVNLGNTC 134
KIM++G E+ + P F E +TP +++ L K LP GI+NLGNTC
Sbjct 67 ICNGSKIMMIGNPEKVVENEAP---VRFYEFMTPQEKSEFLSANKTRRLPCGIMNLGNTC 123
Query 135 YLASVLQMLRPAKDF---SDLLKNYISGGAATAQFAATGQQGALRRLSLALKDFYNQWDQ 191
Y SV Q L P + DLL+ + + Q+ +L+L+L + +Q +
Sbjct 124 YFNSVFQFLLPVSELWESVDLLRAKDATDSDQDQY----------KLALSLAEMRHQLPK 173
Query 192 TVEAVSPFLLVPTLREAYPQFSRRSTSQAGTTGVPMQQDAEECLSCLMT 240
++ P V LR+ P FSR TG+ MQQDAEECLSC+++
Sbjct 174 SISKYVPLAQVQLLRKVNPLFSRTDEK----TGLYMQQDAEECLSCILS 218
> dre:335736 usp14, wu:fk63d09, zgc:55949; ubiquitin specific
protease 14 (tRNA-guanine transglycosylase) (EC:3.4.19.12);
K11843 ubiquitin carboxyl-terminal hydrolase 14 [EC:3.1.2.15]
Length=489
Score = 126 bits (317), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 78/224 (34%), Positives = 125/224 (55%), Gaps = 27/224 (12%)
Query 20 VKWGAKSFN-VQIDPNEPLEVFQAQLYTLTGVPVERQKLLCKGKTIKVDADLQALVSSGC 78
VKWG + F+ V+++ EP VF+AQL+ LTGV ERQK++ KG T+K D + +G
Sbjct 8 VKWGKEKFDAVELNTEEPPMVFKAQLFALTGVQPERQKVMVKGGTLKDDEWGNIKLKNGM 67
Query 79 PKIMLMGTAE---EKTAVKPPPEKTVFVEDLTPAQQAALLKEKKVEPLPNGIVNLGNTCY 135
+++MG+A+ E+ V+P +FVED+T Q A+ ++ LP G+ NLGNTCY
Sbjct 68 -TLLMMGSADALPEEPVVRP-----MFVEDMTEEQLASAME------LPCGLTNLGNTCY 115
Query 136 LASVLQMLRPAKDFSDLLKNYISGGAATAQFAATGQQGALRRLSLALKDFYNQWDQTVEA 195
+ + +Q LR + L+ Y + ++G + ++ AL+D Y D+T +
Sbjct 116 MNATVQCLRSVPELKGALRRY------SGALRSSGANAPSQYITAALRDLYESMDKTSSS 169
Query 196 VSPFLLVPTLREAYPQFSRRSTSQAGTTGVPMQQDAEECLSCLM 239
+ P +L+ L A+PQF+ + G G +QQDA EC +M
Sbjct 170 IPPIILLQFLHMAFPQFAEK-----GDQGQYLQQDANECWVQVM 208
> pfa:PFE1355c ubiquitin carboxyl-terminal hydrolase, putative
(EC:3.1.2.15); K11843 ubiquitin carboxyl-terminal hydrolase
14 [EC:3.1.2.15]
Length=605
Score = 123 bits (309), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 76/226 (33%), Positives = 122/226 (53%), Gaps = 16/226 (7%)
Query 16 VQAAVKWGAKSFN-VQIDPNEPLEVFQAQLYTLTGVPVERQKLLCKGKTIKVDADLQALV 74
V VKW + FN +++D +EPL + + QL+ LT VP E+QKL+ KG +K D DL L
Sbjct 4 VNITVKWKNQVFNNIELDVSEPLILLKTQLWQLTNVPPEKQKLMYKG-LLKDDVDLSLLN 62
Query 75 SSGCPKIMLMGTAEEKTAVKPPPEKTVFVEDLTPAQQAALLKEKKVEPLPNGIVNLGNTC 134
KIML+G+AE ++ P+ +F EDLT ++ + ++ + GIVNLGNTC
Sbjct 63 IKENDKIMLVGSAE---SLVEKPKDIIFEEDLTNEEKQKIHTKENIIFEEQGIVNLGNTC 119
Query 135 YLASVLQMLRPAKDFSDLLKNYISGGAATAQFAATGQQGALRRLSLALKDFYNQWDQTVE 194
Y +VLQ L D + L++Y + ++ T + + +F + ++++ E
Sbjct 120 YFNAVLQFLTSFDDLGNFLRSY---KSKESKLIKTNKDILFD----SFIEFAHSFEKSSE 172
Query 195 AVSPFLLVPTLREAYPQFSRRSTSQAGTTGVPMQQDAEECLSCLMT 240
P L+ + R+ YP+F S T QQDAEEC++ ++T
Sbjct 173 PYVPVTLLKSFRDVYPKFK----SVNLRTKQYAQQDAEECMNAILT 214
> hsa:9097 USP14, TGT; ubiquitin specific peptidase 14 (tRNA-guanine
transglycosylase) (EC:3.4.19.12); K11843 ubiquitin carboxyl-terminal
hydrolase 14 [EC:3.1.2.15]
Length=459
Score = 102 bits (254), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 70/223 (31%), Positives = 105/223 (47%), Gaps = 56/223 (25%)
Query 18 AAVKWGAKSFN-VQIDPNEPLEVFQAQLYTLTGVPVERQKLLCKGKTIKVDADLQALVSS 76
VKWG + F V+++ +EP VF+AQL+ LTGV RQK++ KG T+K D
Sbjct 6 VTVKWGKEKFEGVELNTDEPPMVFKAQLFALTGVQPARQKVMVKGGTLKDD-------DW 58
Query 77 GCPKIMLMGTAEEKTAVKPPPEKTVFVEDLTPAQQAALLKEKKVEPLPNGIVNLGNTCYL 136
G KI M LP G+ NLGNTCY+
Sbjct 59 GNIKIKNM-------------------------------------ELPCGLTNLGNTCYM 81
Query 137 ASVLQMLRPAKDFSDLLKNYISGGAATAQFAATGQQGALRRLSLALKDFYNQWDQTVEAV 196
+ +Q +R + D LK Y GA A+G+ + + ++ AL+D ++ D+T ++
Sbjct 82 NATVQCIRSVPELKDALKRY--AGA----LRASGEMASAQYITAALRDLFDSMDKTSSSI 135
Query 197 SPFLLVPTLREAYPQFSRRSTSQAGTTGVPMQQDAEECLSCLM 239
P +L+ L A+PQF+ + G G +QQDA EC +M
Sbjct 136 PPIILLQFLHMAFPQFAEK-----GEQGQYLQQDANECWIQMM 173
> mmu:59025 Usp14, 2610005K12Rik, 2610037B11Rik, AW107924, C78769,
ax; ubiquitin specific peptidase 14 (EC:3.4.19.12); K11843
ubiquitin carboxyl-terminal hydrolase 14 [EC:3.1.2.15]
Length=458
Score = 102 bits (254), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 69/221 (31%), Positives = 106/221 (47%), Gaps = 56/221 (25%)
Query 20 VKWGAKSFN-VQIDPNEPLEVFQAQLYTLTGVPVERQKLLCKGKTIKVDADLQALVSSGC 78
VKWG + F V+++ +EP VF+AQL+ LTGV RQK++ KG T+K D
Sbjct 8 VKWGKEKFEGVELNTDEPPMVFKAQLFALTGVQPARQKVMVKGGTLKDD----------- 56
Query 79 PKIMLMGTAEEKTAVKPPPEKTVFVEDLTPAQQAALLKEKKVEPLPNGIVNLGNTCYLAS 138
+K K +E LP G+ NLGNTCY+ +
Sbjct 57 --------------------------------DWGNIKMKNME-LPCGLTNLGNTCYMNA 83
Query 139 VLQMLRPAKDFSDLLKNYISGGAATAQFAATGQQGALRRLSLALKDFYNQWDQTVEAVSP 198
+Q +R + D LK Y GA A+G+ + + ++ AL+D ++ D+T ++ P
Sbjct 84 TVQCIRSVPELKDALKRY--AGA----LRASGEMASAQYITAALRDLFDSMDKTSSSIPP 137
Query 199 FLLVPTLREAYPQFSRRSTSQAGTTGVPMQQDAEECLSCLM 239
+L+ L A+PQF+ + G G +QQDA EC +M
Sbjct 138 IILLQFLHMAFPQFAEK-----GEQGQYLQQDANECWIQMM 173
> sce:YFR010W UBP6; Ubiquitin-specific protease situated in the
base subcomplex of the 26S proteasome, releases free ubiquitin
from branched polyubiquitin chains; works in opposition
to polyubiquitin elongation activity of Hul5p (EC:3.1.2.15);
K11843 ubiquitin carboxyl-terminal hydrolase 14 [EC:3.1.2.15]
Length=499
Score = 93.6 bits (231), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 67/200 (33%), Positives = 103/200 (51%), Gaps = 25/200 (12%)
Query 47 LTGVPVERQKLLCKG-----KTIKVDADLQALVSSGCPKIMLMGTAEEKTAVKPPPEKTV 101
LT VP RQK + KG ++IK+ L+ G +ML+GT + + P +K
Sbjct 37 LTQVPSARQKYMVKGGLSGEESIKI----YPLIKPGST-VMLLGTPD-ANLISKPAKKNN 90
Query 102 FVEDLTPAQQAALLKEKKVEPLPNGIVNLGNTCYLASVLQMLRPAKDFSDLLKNYI-SGG 160
F+EDL P QQ ++ LP G N+GNTCYL + LQ L D D++ NY S G
Sbjct 91 FIEDLAPEQQV-----QQFAQLPVGFKNMGNTCYLNATLQALYRVNDLRDMILNYNPSQG 145
Query 161 AATAQFAATGQQGALRRLSLALKD-FYNQWDQTVEAVSPFLLVPTLREAYPQFSRRSTSQ 219
+ + + +++ + +K F N +++ ++V P +L+ TLR+ YPQF+ R +
Sbjct 146 VSN---SGAQDEEIHKQIVIEMKRCFENLQNKSFKSVLPIVLLNTLRKCYPQFAERDSQ- 201
Query 220 AGTTGVPMQQDAEECLSCLM 239
G QQDAEE + L
Sbjct 202 ---GGFYKQQDAEELFTQLF 218
> ath:AT1G51710 UBP6; UBP6 (UBIQUITIN-SPECIFIC PROTEASE 6); calmodulin
binding / ubiquitin-specific protease; K11843 ubiquitin
carboxyl-terminal hydrolase 14 [EC:3.1.2.15]
Length=443
Score = 92.8 bits (229), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 63/173 (36%), Positives = 87/173 (50%), Gaps = 20/173 (11%)
Query 67 DADLQALVSSGCPKIMLMGTAEEKTAVKPPPEKTVFVEDLTPAQQAALLKEKKVEPLPNG 126
D D A+ K+M+MGTA+E VK P + VF EDL A L G
Sbjct 15 DGDWAAIGVKDGQKLMMMGTADE--IVKAPEKAIVFAEDLPEEALATNL------GYSAG 66
Query 127 IVNLGNTCYLASVLQMLRPAKDFSDLLKNYISGGAATAQFAATGQQGALRRLSLALKDFY 186
+VNLGNTCY+ S +Q L+ + L NY S A + T L++A ++ +
Sbjct 67 LVNLGNTCYMNSTVQCLKSVPELKSALSNY-SLAARSNDVDQTSHM-----LTVATRELF 120
Query 187 NQWDQTVEAVSPFLLVPTLREAYPQFSRRSTSQAGTTGVPMQQDAEECLSCLM 239
+ D++V AVSP LR+ YPQFS+ G+ MQQDAEEC + L+
Sbjct 121 GELDRSVNAVSPSQFWMVLRKKYPQFSQLQ------NGMHMQQDAEECWTQLL 167
> cel:C13B4.2 usp-14; Ubiquitin Specific Protease family member
(usp-14); K11843 ubiquitin carboxyl-terminal hydrolase 14
[EC:3.1.2.15]
Length=489
Score = 84.7 bits (208), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 69/225 (30%), Positives = 103/225 (45%), Gaps = 33/225 (14%)
Query 20 VKWGAKSFNVQIDPNEPLEVFQAQLYTLTGVPVERQKLLCKGKTIKVDADLQALVSSGCP 79
VKW + + V++D + P VF+AQL+ LT V ERQK++ G+T+ D D + +
Sbjct 6 VKWQKEKYVVEVDTSAPPMVFKAQLFALTQVVPERQKVVIMGRTLG-DDDWEGITIKENM 64
Query 80 KIMLMGTAEEKTAVKPPP--EKTVFVEDLTPAQQAALLKEKKVEPLPNGIVNLGNTCYLA 137
IM+MG+ E KPP EK D + +AL P G+ NLGNTCY
Sbjct 65 TIMMMGSVGE--IPKPPTVLEKKQANRDKQAEEISAL--------YPCGLANLGNTCYFN 114
Query 138 SVLQMLRPAKDFSDLLKNYISGGAATAQFAATGQQ--GALRRLSLALKDFYNQWDQTVEA 195
S +QML+ + + A + + L L +L+D E
Sbjct 115 SCVQMLKEVNEL-------VLKPAEEMRIREHNDRLCHNLATLFNSLRDKDRALRSKGEP 167
Query 196 VSPFLLVPTLREAYPQFSRRSTSQAGTTGVPMQQDAEECLSCLMT 240
+ PF + TL +++PQF + QQDA ECL +M+
Sbjct 168 IKPFAAILTLSDSFPQFEKFK-----------QQDANECLVSIMS 201
> mmu:79560 Ublcp1, 4930527B16Rik, 8430435I17Rik, BC002236, MGC117826,
MGC7513; ubiquitin-like domain containing CTD phosphatase
1 (EC:3.1.3.16); K01090 protein phosphatase [EC:3.1.3.16]
Length=318
Score = 50.1 bits (118), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 36/93 (38%), Positives = 50/93 (53%), Gaps = 6/93 (6%)
Query 20 VKWGAKSFNVQ-IDPNEPLEVFQAQLYTLTGVPVERQKLL---CKGKTIKVDADLQALVS 75
VKWG + ++V + ++ + + L TLTGV ERQKLL KGK + D L AL
Sbjct 7 VKWGGQEYSVTTLSEDDTVLDLKQFLKTLTGVLPERQKLLGLKVKGKPAENDVKLGALKL 66
Query 76 SGCPKIMLMGTAEE--KTAVKPPPEKTVFVEDL 106
KIM+MGT EE + + PPP+ + D
Sbjct 67 KPNTKIMMMGTREESLEDVLCPPPDNDDVINDF 99
> mmu:100045709 ubiquitin-like domain-containing CTD phosphatase
1-like
Length=310
Score = 50.1 bits (118), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 36/93 (38%), Positives = 50/93 (53%), Gaps = 6/93 (6%)
Query 20 VKWGAKSFNVQ-IDPNEPLEVFQAQLYTLTGVPVERQKLL---CKGKTIKVDADLQALVS 75
VKWG + ++V + ++ + + L TLTGV ERQKLL KGK + D L AL
Sbjct 7 VKWGGQEYSVTTLSEDDTVLDLKQFLKTLTGVLPERQKLLGLKVKGKPAENDVKLGALKL 66
Query 76 SGCPKIMLMGTAEE--KTAVKPPPEKTVFVEDL 106
KIM+MGT EE + + PPP+ + D
Sbjct 67 KPNTKIMMMGTREESLEDVLCPPPDNDDVINDF 99
> hsa:134510 UBLCP1, CPUB1, FLJ25267, MGC10067; ubiquitin-like
domain containing CTD phosphatase 1 (EC:3.1.3.16); K01090 protein
phosphatase [EC:3.1.3.16]
Length=318
Score = 48.5 bits (114), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 35/84 (41%), Positives = 47/84 (55%), Gaps = 6/84 (7%)
Query 20 VKWGAKSFNVQ-IDPNEPLEVFQAQLYTLTGVPVERQKLL---CKGKTIKVDADLQALVS 75
VKWG + ++V + ++ + + L TLTGV ERQKLL KGK + D L AL
Sbjct 7 VKWGGQEYSVTTLSEDDTVLDLKQFLKTLTGVLPERQKLLGLKVKGKPAENDVKLGALKL 66
Query 76 SGCPKIMLMGTAEE--KTAVKPPP 97
KIM+MGT EE + + PPP
Sbjct 67 KPNTKIMMMGTREESLEDVLGPPP 90
> xla:447721 ublcp1; ubiquitin-like domain containing CTD phosphatase
1 (EC:3.1.3.16); K01090 protein phosphatase [EC:3.1.3.16]
Length=318
Score = 45.4 bits (106), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 33/93 (35%), Positives = 46/93 (49%), Gaps = 6/93 (6%)
Query 20 VKWGAKSFNVQ-IDPNEPLEVFQAQLYTLTGVPVERQKLL---CKGKTIKVDADLQALVS 75
+KWG + F + + + + + L +LTGV ER KLL KGK + D L L
Sbjct 7 IKWGGQEFPLSALSEEDTVLDLKHSLKSLTGVLPERMKLLGLKYKGKPAENDVKLGVLRL 66
Query 76 SGCPKIMLMGTAEE--KTAVKPPPEKTVFVEDL 106
KIM+MGT EE + + PPP+ V D
Sbjct 67 KPNTKIMMMGTREESLEEMMAPPPDNDEVVNDF 99
> mmu:30941 Usp21, ESTM28, Usp16, Usp23, W53272; ubiquitin specific
peptidase 21 (EC:3.4.19.12); K11833 ubiquitin carboxyl-terminal
hydrolase 2/21 [EC:3.1.2.15]
Length=566
Score = 44.3 bits (103), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 49/148 (33%), Positives = 59/148 (39%), Gaps = 41/148 (27%)
Query 102 FVEDLTPAQQAALLKEKKVEPLPNGIVNLGNTCYLASVLQML---RPAKDF---SDLLKN 155
F D A LL V G+ NLGNTC+L +VLQ L RP +DF D +
Sbjct 194 FYSDDKMAHHTLLLGSGHV-----GLRNLGNTCFLNAVLQCLSSTRPLRDFCLRRDFRQE 248
Query 156 YISGGAA---TAQFAATGQQGALRRLSLALKDFYNQWD-QTVEAVSPFLLVPTLREAYPQ 211
GG A T FA GAL W + EAV+P ++ P
Sbjct 249 VPGGGRAQELTEAFADV--IGAL-------------WHPDSCEAVNPTRFRAVFQKYVPS 293
Query 212 FSRRSTSQAGTTGVPMQQDAEECLSCLM 239
FS S QQDA+E L LM
Sbjct 294 FSGYS-----------QQDAQEFLKLLM 310
> hsa:27005 USP21, MGC3394, USP16, USP23; ubiquitin specific peptidase
21 (EC:3.4.19.12); K11833 ubiquitin carboxyl-terminal
hydrolase 2/21 [EC:3.1.2.15]
Length=565
Score = 44.3 bits (103), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 49/148 (33%), Positives = 59/148 (39%), Gaps = 41/148 (27%)
Query 102 FVEDLTPAQQAALLKEKKVEPLPNGIVNLGNTCYLASVLQML---RPAKDF---SDLLKN 155
F D A LL V G+ NLGNTC+L +VLQ L RP +DF D +
Sbjct 194 FYSDDKMAHHTLLLGSGHV-----GLRNLGNTCFLNAVLQCLSSTRPLRDFCLRRDFRQE 248
Query 156 YISGGAA---TAQFAATGQQGALRRLSLALKDFYNQWD-QTVEAVSPFLLVPTLREAYPQ 211
GG A T FA GAL W + EAV+P ++ P
Sbjct 249 VPGGGRAQELTEAFADV--IGAL-------------WHPDSCEAVNPTRFRAVFQKYVPS 293
Query 212 FSRRSTSQAGTTGVPMQQDAEECLSCLM 239
FS S QQDA+E L LM
Sbjct 294 FSGYS-----------QQDAQEFLKLLM 310
> dre:559473 usp49, si:rp71-11b17.7; ubiquitin specific peptidase
49 (EC:3.1.2.15); K11834 ubiquitin carboxyl-terminal hydrolase
44/49 [EC:3.1.2.15]
Length=649
Score = 43.9 bits (102), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 39/75 (52%), Gaps = 5/75 (6%)
Query 79 PKIMLMGTAEEKTAVKPPPEKTVFVEDLTPAQQAALLKEKKVEPLPNGIVNLGNTCYLAS 138
PK +L T KT + P +T + +Q A + +K+ P G+ NLGNTCY+ S
Sbjct 224 PKQLL--TLSAKTLSRKPLRRTARLHRYHASQSA---RRRKLAPGVTGLRNLGNTCYMNS 278
Query 139 VLQMLRPAKDFSDLL 153
+LQ+L + F +
Sbjct 279 ILQVLSHLQKFRECF 293
> tpv:TP01_0316 ubiquitin carboxyl-terminal hydrolase a; K11836
ubiquitin carboxyl-terminal hydrolase 5/13 [EC:3.1.2.15]
Length=777
Score = 43.9 bits (102), Expect = 5e-04, Method: Composition-based stats.
Identities = 23/51 (45%), Positives = 31/51 (60%), Gaps = 3/51 (5%)
Query 106 LTPAQQAALLKEKKVEPLPNGIVNLGNTCYLASVLQMLRPAKDFSDLLKNY 156
L+ A A+L+ ++P GI+NLGN CYL SVLQML K+ +D Y
Sbjct 294 LSDASGASLIH---LQPKLIGILNLGNNCYLNSVLQMLNNVKELNDFFLQY 341
> cel:F07A11.4 hypothetical protein; K11847 ubiquitin carboxyl-terminal
hydrolase 19 [EC:3.1.2.15]
Length=1095
Score = 42.7 bits (99), Expect = 0.001, Method: Composition-based stats.
Identities = 36/121 (29%), Positives = 54/121 (44%), Gaps = 16/121 (13%)
Query 120 VEPLPNGIVNLGNTCYLASVLQMLRPAKDFSDLLKNYISGGAATAQFAATGQQGALRRLS 179
VE G+ N+GNTC++ +VLQML + L+ Y + T G+ RL+
Sbjct 418 VEQGYTGLRNIGNTCFMNAVLQMLVNNIE----LREYFLRNHYQPEINETNPLGSEGRLA 473
Query 180 LALKDFYNQ-WDQTVEAVSPFLLVPTLREAYPQFSRRSTSQAGTTGVPMQQDAEECLSCL 238
A DF +Q W +A+ P + + E QF+ + Q DA E LS L
Sbjct 474 KAFADFMHQMWSGHQKAIEPTQIKNIVAEKASQFANFA-----------QHDAHEFLSFL 522
Query 239 M 239
+
Sbjct 523 L 523
> dre:561425 novel protein similar to vertebrate ubiquitin specific
peptidase 19 (USP19); K11847 ubiquitin carboxyl-terminal
hydrolase 19 [EC:3.1.2.15]
Length=1489
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 40/150 (26%), Positives = 65/150 (43%), Gaps = 22/150 (14%)
Query 97 PEKTVFVEDLTPA-----QQAALLKEKKV-EPLPNGIVNLGNTCYLASVLQMLRPAKDFS 150
P+ T V+ +T +++ ++EKKV P G+VNLGNTC++ SV+Q L ++
Sbjct 604 PKPTCMVQPMTHTPPVGNERSEEVEEKKVCHPGFTGLVNLGNTCFMNSVIQSLSNTRE-- 661
Query 151 DLLKNYISGGAATAQFAATGQQGALRRLSLALKDFYNQ-WDQTVEAVSPFLLVPTLREAY 209
L++Y ++ G RL++ W T A P L +
Sbjct 662 --LRDYFHDRGFESEINCNNPLGTGGRLAIGFAVLLRALWKGTHHAFQPSKLKAIVASKA 719
Query 210 PQFSRRSTSQAGTTGVPMQQDAEECLSCLM 239
QF TG Q DA+E ++ L+
Sbjct 720 SQF----------TGY-AQHDAQEFMAFLL 738
> ath:AT2G30100 ubiquitin family protein
Length=897
Score = 40.8 bits (94), Expect = 0.004, Method: Composition-based stats.
Identities = 28/82 (34%), Positives = 45/82 (54%), Gaps = 2/82 (2%)
Query 6 LSPQKMKASEVQAAVKWGAKSFNVQIDPNEPLEVFQAQLYTLTGVPVERQKLLCKGKTIK 65
L M S ++ VK+G KS + + P+ ++ ++QL +T V QKL+ KGK +
Sbjct 526 LEDPTMADSTIKLTVKFGGKSIPLSVSPDCTVKDLKSQLQPITNVLPRGQKLIFKGKVLV 585
Query 66 VDADL-QALVSSGCPKIMLMGT 86
+ L Q+ V SG K+MLM +
Sbjct 586 ETSTLKQSDVGSGA-KLMLMAS 606
> cel:K09A9.4 hypothetical protein
Length=716
Score = 40.4 bits (93), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 30/102 (29%), Positives = 49/102 (48%), Gaps = 14/102 (13%)
Query 118 KKVEPLPN----GIVNLGNTCYLASVLQMLRPAKDFSDLLKNYISGGA---ATAQFAATG 170
+K+EPL G +N GNTCY+ +VLQ+L F+ L I G + T
Sbjct 171 RKMEPLAFRGLLGYLNFGNTCYMNAVLQLLGHCSPFTQYLIELIPPGGWSNCSHDIPKTA 230
Query 171 QQGALRRLSLALKDFYNQWDQTVEAVSPFLLVPTLREAYPQF 212
Q ++L L++ Y+ D + +SP+ ++ +R P F
Sbjct 231 IQ-----MALDLRNMYS--DFPLPYLSPWKIITCVRNEMPGF 265
> mmu:75083 Usp50, 1700086G18Rik, 4930511O11Rik; ubiquitin specific
peptidase 50 (EC:3.4.19.12)
Length=390
Score = 40.0 bits (92), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 35/124 (28%), Positives = 55/124 (44%), Gaps = 22/124 (17%)
Query 115 LKEKKVEPLPNGIV---NLGNTCYLASVLQMLRPAKDFSDLLKNYISGGAATAQFAATGQ 171
L E K +P G+ NLGNTCY+ ++LQ L S L++ ++SG TA +
Sbjct 32 LPESKTQPHFQGVTGLRNLGNTCYMNAILQCLCSV---SPLVEYFLSGKYITALKKDCSE 88
Query 172 QGALRRLSLALKDFYNQWDQTVEAVSPFLLVPTLREAYPQFSRRSTSQAGTTGVPMQQDA 231
+ + D W + VSP + + + YP F +++ QQDA
Sbjct 89 --VTTAFAYLMTD---MWLGDSDCVSPEIFLSAVGSLYPAFLKKT-----------QQDA 132
Query 232 EECL 235
+E L
Sbjct 133 QEFL 136
> mmu:230484 Usp1, MGC25559; ubiquitin specific peptidase 1 (EC:3.4.19.12);
K11832 ubiquitin carboxyl-terminal hydrolase 1
[EC:3.1.2.15]
Length=784
Score = 39.7 bits (91), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 52/178 (29%), Positives = 77/178 (43%), Gaps = 41/178 (23%)
Query 88 EEKTAVKPPPEKTVFVEDLTPAQQAALLK-EKKVEPLPN-GIVNLGNTCYLASVLQMLRP 145
EEKT+ E ++ + PA Q++ + EK+ LP G+ NLGNTCYL S+LQ+L
Sbjct 46 EEKTSEYRGSE----IDQVVPAAQSSPVSCEKRENLLPFVGLNNLGNTCYLNSILQVLYF 101
Query 146 AKDFSDLLK---NYIS-GGAATAQFAATGQQGALRRLSLALKDFYNQWDQTVEAV----S 197
F +K N IS A + +G+ + SLA + + +V +
Sbjct 102 CPGFKTGVKHLFNIISRKKEALKDDSNQKDKGSCKEESLASYELICSLQSLIISVEQLQA 161
Query 198 PFLLVP----------------TLREAYPQFSRRSTSQAGTTGVPMQQDAEECLSCLM 239
FLL P TLRE P + G +Q DA+E L C++
Sbjct 162 SFLLNPEKYTDELATQPRRLLNTLRELNPMYE----------GF-LQHDAQEVLQCIL 208
> ath:AT5G25270 hypothetical protein
Length=658
Score = 39.7 bits (91), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 33/49 (67%), Gaps = 0/49 (0%)
Query 24 AKSFNVQIDPNEPLEVFQAQLYTLTGVPVERQKLLCKGKTIKVDADLQA 72
++++ +++D P+ + Q+ ++TGV E+Q+L+C+GK +K D L A
Sbjct 30 SQTYTLRVDKCVPVPALKEQVASVTGVVTEQQRLICRGKVMKDDQLLSA 78
> mmu:72344 Usp36, 2700002L06Rik, mKIAA1453; ubiquitin specific
peptidase 36 (EC:3.1.2.15 3.4.19.12); K11855 ubiquitin carboxyl-terminal
hydrolase 36/42 [EC:3.1.2.15]
Length=1098
Score = 39.7 bits (91), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 32/119 (26%), Positives = 54/119 (45%), Gaps = 16/119 (13%)
Query 95 PPPEKTVF-VEDLTPAQQAALLKEKKVEPLPNGIVNLGNTCYLASVLQMLRPAKDFSDLL 153
P P+K +F VE L+ L+ ++V + G+ NLGNTC+L S +Q L ++ L
Sbjct 98 PAPQKVLFPVERLS-------LRWERVFRVGAGLHNLGNTCFLNSTIQCL----TYTPPL 146
Query 154 KNYISGGAATAQFAATGQQGALRRLSLALKDFYNQWDQTVEAVSPFLLVPTLREAYPQF 212
NY+ + + A + QG L L + + A+ P + L++ F
Sbjct 147 ANYL----LSKEHARSCHQGGFCMLCLMQNHMVQAFANSGNAIKPVSFIRDLKKIARHF 201
> xla:447469 usp38, MGC81730; ubiquitin specific peptidase 38
(EC:3.1.2.15); K11854 ubiquitin carboxyl-terminal hydrolase
35/38 [EC:3.1.2.15]
Length=1037
Score = 39.3 bits (90), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 18/31 (58%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 119 KVEPLPNGIVNLGNTCYLASVLQMLRPAKDF 149
K E G++NLGNTCY+ SVLQ L A DF
Sbjct 438 KSETGKTGLINLGNTCYMNSVLQALFMATDF 468
> ath:AT5G42220 ubiquitin family protein
Length=879
Score = 39.3 bits (90), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 33/47 (70%), Gaps = 0/47 (0%)
Query 24 AKSFNVQIDPNEPLEVFQAQLYTLTGVPVERQKLLCKGKTIKVDADL 70
++++ Q++ NE + +F+ ++ + TGVPV +Q+L+ +G+ +K D L
Sbjct 33 SRTYTFQVNKNETVLLFKEKIASETGVPVGQQRLIFRGRVLKDDHPL 79
> hsa:25862 USP49, MGC20741; ubiquitin specific peptidase 49 (EC:3.4.19.12);
K11834 ubiquitin carboxyl-terminal hydrolase
44/49 [EC:3.1.2.15]
Length=640
Score = 38.9 bits (89), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 27/48 (56%), Gaps = 0/48 (0%)
Query 108 PAQQAALLKEKKVEPLPNGIVNLGNTCYLASVLQMLRPAKDFSDLLKN 155
PA L ++ + P G+ NLGNTCY+ S+LQ+L + F + N
Sbjct 236 PAATLKLRRQPAMAPGVTGLRNLGNTCYMNSILQVLSHLQKFRECFLN 283
> hsa:7398 USP1, UBP; ubiquitin specific peptidase 1 (EC:3.4.19.12);
K11832 ubiquitin carboxyl-terminal hydrolase 1 [EC:3.1.2.15]
Length=785
Score = 38.5 bits (88), Expect = 0.020, Method: Composition-based stats.
Identities = 47/163 (28%), Positives = 69/163 (42%), Gaps = 37/163 (22%)
Query 103 VEDLTPAQQAALLK-EKKVEPLP-NGIVNLGNTCYLASVLQMLRPAKDFSDLLK---NYI 157
++ + PA Q++ + EK+ LP G+ NLGNTCYL S+LQ+L F +K N I
Sbjct 57 IDQVVPAAQSSPINCEKRENLLPFVGLNNLGNTCYLNSILQVLYFCPGFKSGVKHLFNII 116
Query 158 S-GGAATAQFAATGQQGALRRLSLALKDFYNQWDQTVEAV----SPFLLVP--------- 203
S A A +G + SLA + + +V + FLL P
Sbjct 117 SRKKEALKDEANQKDKGNCKEDSLASYELICSLQSLIISVEQLQASFLLNPEKYTDELAT 176
Query 204 -------TLREAYPQFSRRSTSQAGTTGVPMQQDAEECLSCLM 239
TLRE P + +Q DA+E L C++
Sbjct 177 QPRRLLNTLRELNPMYEGY-----------LQHDAQEVLQCIL 208
> mmu:100045226 ubiquitin carboxyl-terminal hydrolase 35-like
Length=714
Score = 38.5 bits (88), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 16/24 (66%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 126 GIVNLGNTCYLASVLQMLRPAKDF 149
G++NLGNTCY+ SVLQ L A DF
Sbjct 205 GLINLGNTCYVNSVLQALFMASDF 228
> dre:322007 usp1, wu:fb43a02, zgc:55962; ubiquitin specific protease
1 (EC:3.4.19.12); K11832 ubiquitin carboxyl-terminal
hydrolase 1 [EC:3.1.2.15]
Length=772
Score = 38.5 bits (88), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 40/71 (56%), Gaps = 3/71 (4%)
Query 88 EEKTAVKPPPEKTVFVEDLTP--AQQAALLKEKKVEPLPN-GIVNLGNTCYLASVLQMLR 144
E+ + V P++T+ + + P + L EK+ +P G+ NLGNTCYL S+LQ+L
Sbjct 45 EDNSNVTTEPQETICHDQVVPLPCPSSPLTCEKRESLVPFVGLNNLGNTCYLNSILQVLY 104
Query 145 PAKDFSDLLKN 155
F + +K+
Sbjct 105 YCPGFKEAIKS 115
> hsa:84640 USP38, FLJ35970, HP43.8KD, KIAA1891; ubiquitin specific
peptidase 38 (EC:3.4.19.12); K11854 ubiquitin carboxyl-terminal
hydrolase 35/38 [EC:3.1.2.15]
Length=1042
Score = 38.5 bits (88), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 17/31 (54%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 119 KVEPLPNGIVNLGNTCYLASVLQMLRPAKDF 149
K E G++NLGNTCY+ SV+Q L A DF
Sbjct 439 KSETGKTGLINLGNTCYMNSVIQALFMATDF 469
> mmu:244144 Usp35, Gm1088, Gm493; ubiquitin specific peptidase
35; K11854 ubiquitin carboxyl-terminal hydrolase 35/38 [EC:3.1.2.15]
Length=1009
Score = 38.5 bits (88), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 16/24 (66%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 126 GIVNLGNTCYLASVLQMLRPAKDF 149
G++NLGNTCY+ SVLQ L A DF
Sbjct 442 GLINLGNTCYVNSVLQALFMASDF 465
> ath:AT3G14400 UBP25; UBP25 (UBIQUITIN-SPECIFIC PROTEASE 25);
ubiquitin thiolesterase/ ubiquitin-specific protease
Length=661
Score = 38.1 bits (87), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 18/31 (58%), Positives = 22/31 (70%), Gaps = 0/31 (0%)
Query 113 ALLKEKKVEPLPNGIVNLGNTCYLASVLQML 143
+LL +K+ P G+ NLGNTCYL SVLQ L
Sbjct 12 SLLSQKRRNGPPLGLRNLGNTCYLNSVLQCL 42
> pfa:PFD0680c ubiquitin carboxyl-terminal hydrolase A, putative
(EC:3.1.2.15); K11836 ubiquitin carboxyl-terminal hydrolase
5/13 [EC:3.1.2.15]
Length=853
Score = 38.1 bits (87), Expect = 0.027, Method: Composition-based stats.
Identities = 19/33 (57%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 126 GIVNLGNTCYLASVLQMLRPAKDFSDLLKNYIS 158
G VNLGNTCY+ S LQ+L KD S N IS
Sbjct 324 GFVNLGNTCYMNSALQVLLSIKDISYRYLNNIS 356
> dre:557981 ubiquitin specific peptidase 15-like; K11837 ubiquitin
carboxyl-terminal hydrolase 6/32 [EC:3.1.2.15]
Length=1675
Score = 38.1 bits (87), Expect = 0.029, Method: Composition-based stats.
Identities = 34/144 (23%), Positives = 61/144 (42%), Gaps = 20/144 (13%)
Query 97 PEKTVFVEDLTPAQQAALLKEKKVEPLPNGIVNLGNTCYLASVLQMLRPAKDFSDLLKNY 156
PE+ F+ + + + + EK G+ NLGNTC++ S +Q + K ++ Y
Sbjct 800 PEEMSFIANSSKIDRHKVPTEKGA----TGLSNLGNTCFMNSSIQCVSNTKPLTE----Y 851
Query 157 ISGGAATAQFAATGQQGALRRLSLALKDFYNQ-WDQTVEAVSPFLLVPTLREAYPQFSRR 215
G + T G ++ D + W T + V+P L T+ + P+F+
Sbjct 852 FISGRHLYELNRTNPIGMRGHMAKCYGDLVQELWSGTQKNVAPLKLRWTIAKYAPRFNGF 911
Query 216 STSQAGTTGVPMQQDAEECLSCLM 239
QQD++E L+ L+
Sbjct 912 Q-----------QQDSQELLAFLL 924
> mmu:224836 Usp49, C330046L10Rik, Gm545; ubiquitin specific peptidase
49 (EC:3.4.19.12); K11834 ubiquitin carboxyl-terminal
hydrolase 44/49 [EC:3.1.2.15]
Length=685
Score = 38.1 bits (87), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 27/50 (54%), Gaps = 6/50 (12%)
Query 106 LTPAQQAALLKEKKVEPLPNGIVNLGNTCYLASVLQMLRPAKDFSDLLKN 155
L P +Q A V P G+ NLGNTCY+ S+LQ+L + F + N
Sbjct 237 LNPRRQPA------VAPGVTGLRNLGNTCYMNSILQVLSHLQKFRECFLN 280
> mmu:71472 Usp19, 8430421I07Rik, AI047774, Zmynd9; ubiquitin
specific peptidase 19 (EC:3.4.19.12); K11847 ubiquitin carboxyl-terminal
hydrolase 19 [EC:3.1.2.15]
Length=1324
Score = 37.7 bits (86), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 31/115 (26%), Positives = 50/115 (43%), Gaps = 16/115 (13%)
Query 126 GIVNLGNTCYLASVLQMLRPAKDFSDLLKNYISGGAATAQFAATGQQGALRRLSLALKDF 185
G+VNLGNTC++ SV+Q L ++ L+++ + A+ G RL++
Sbjct 541 GLVNLGNTCFMNSVIQSLSNTRE----LRDFFHDRSFEAEINYNNPLGTGGRLAIGFAVL 596
Query 186 YNQ-WDQTVEAVSPFLLVPTLREAYPQFSRRSTSQAGTTGVPMQQDAEECLSCLM 239
W T +A P L + QF TG Q DA+E ++ L+
Sbjct 597 LRALWKGTHQAFQPSKLKAIVASKASQF----------TGY-AQHDAQEFMAFLL 640
> sce:YER151C UBP3, BLM3; Ubiquitin-specific protease that interacts
with Bre5p to co-regulate anterograde and retrograde
transport between endoplasmic reticulum and Golgi compartments;
inhibitor of gene silencing; cleaves ubiquitin fusions but
not polyubiquitin (EC:3.1.2.15); K11841 ubiquitin carboxyl-terminal
hydrolase 10 [EC:3.1.2.15]
Length=912
Score = 37.7 bits (86), Expect = 0.034, Method: Composition-based stats.
Identities = 36/129 (27%), Positives = 56/129 (43%), Gaps = 18/129 (13%)
Query 41 QAQLYTLTGVPVERQKLLCKGKTIKVDADLQALVSSGCPKIMLMGTAEEKTAVKPPPEKT 100
QA T++G V + + G T V + A + G L+ +K K P T
Sbjct 362 QASNKTVSGSMVTKTPI--SGTTAGVSSTNMAAATIGKSSSPLLSKQPQKKDKKYVPPST 419
Query 101 VFVEDL---------TPAQQAALLKEKKVEP-------LPNGIVNLGNTCYLASVLQMLR 144
+E L P + +L+ K VE +P GI+N N C+++SVLQ+L
Sbjct 420 KGIEPLGSIALRMCFDPDFISYVLRNKDVENKIPVHSIIPRGIINRANICFMSSVLQVLL 479
Query 145 PAKDFSDLL 153
K F D++
Sbjct 480 YCKPFIDVI 488
> hsa:57558 USP35; ubiquitin specific peptidase 35 (EC:3.4.19.12);
K11854 ubiquitin carboxyl-terminal hydrolase 35/38 [EC:3.1.2.15]
Length=1018
Score = 37.7 bits (86), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 15/24 (62%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 126 GIVNLGNTCYLASVLQMLRPAKDF 149
G++NLGNTCY+ S+LQ L A DF
Sbjct 442 GLINLGNTCYVNSILQALFMASDF 465
> mmu:237898 Usp32, 2900074J03Rik, 6430526O11Rik, AW045245; ubiquitin
specific peptidase 32 (EC:3.1.2.15); K11837 ubiquitin
carboxyl-terminal hydrolase 6/32 [EC:3.1.2.15]
Length=1604
Score = 37.4 bits (85), Expect = 0.044, Method: Composition-based stats.
Identities = 34/144 (23%), Positives = 60/144 (41%), Gaps = 20/144 (13%)
Query 97 PEKTVFVEDLTPAQQAALLKEKKVEPLPNGIVNLGNTCYLASVLQMLRPAKDFSDLLKNY 156
PE+ F+ + + + + EK G+ NLGNTC++ S +Q + + L Y
Sbjct 710 PEEMSFIANSSKIDRHKVPTEKGA----TGLSNLGNTCFMNSSIQCVSNTQP----LTQY 761
Query 157 ISGGAATAQFAATGQQGALRRLSLALKDFYNQ-WDQTVEAVSPFLLVPTLREAYPQFSRR 215
G + T G ++ D + W T + V+P L T+ + P+F+
Sbjct 762 FISGRHLYELNRTNPIGMKGHMAKCYGDLVQELWSGTQKNVAPLKLRWTIAKYAPRFNGF 821
Query 216 STSQAGTTGVPMQQDAEECLSCLM 239
QQD++E L+ L+
Sbjct 822 Q-----------QQDSQELLAFLL 834
> dre:559099 usp30, si:dkey-72n1.2; ubiquitin specific peptidase
30 (EC:3.4.19.12); K11851 ubiquitin carboxyl-terminal hydrolase
30 [EC:3.1.2.15]
Length=491
Score = 37.4 bits (85), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 29/47 (61%), Gaps = 0/47 (0%)
Query 116 KEKKVEPLPNGIVNLGNTCYLASVLQMLRPAKDFSDLLKNYISGGAA 162
+ KK + + G++NLGNTC++ S+LQ L F L+++ S +A
Sbjct 55 RRKKRKGMVPGLLNLGNTCFMNSLLQGLAACPSFIRWLEDFTSQNSA 101
> hsa:10869 USP19, ZMYND9; ubiquitin specific peptidase 19 (EC:3.4.19.12);
K11847 ubiquitin carboxyl-terminal hydrolase 19
[EC:3.1.2.15]
Length=1419
Score = 37.4 bits (85), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 31/115 (26%), Positives = 49/115 (42%), Gaps = 16/115 (13%)
Query 126 GIVNLGNTCYLASVLQMLRPAKDFSDLLKNYISGGAATAQFAATGQQGALRRLSLALKDF 185
G+VNLGNTC++ SV+Q L ++ L+++ + A+ G RL++
Sbjct 599 GLVNLGNTCFMNSVIQSLSNTRE----LRDFFHDRSFEAEINYNNPLGTGGRLAIGFAVL 654
Query 186 YNQ-WDQTVEAVSPFLLVPTLREAYPQFSRRSTSQAGTTGVPMQQDAEECLSCLM 239
W T A P L + QF TG Q DA+E ++ L+
Sbjct 655 LRALWKGTHHAFQPSKLKAIVASKASQF----------TGY-AQHDAQEFMAFLL 698
> hsa:84669 USP32, NY-REN-60, USP10; ubiquitin specific peptidase
32 (EC:3.4.19.12); K11837 ubiquitin carboxyl-terminal hydrolase
6/32 [EC:3.1.2.15]
Length=1604
Score = 37.4 bits (85), Expect = 0.054, Method: Composition-based stats.
Identities = 34/144 (23%), Positives = 60/144 (41%), Gaps = 20/144 (13%)
Query 97 PEKTVFVEDLTPAQQAALLKEKKVEPLPNGIVNLGNTCYLASVLQMLRPAKDFSDLLKNY 156
PE+ F+ + + + + EK G+ NLGNTC++ S +Q + + L Y
Sbjct 710 PEEMSFIANSSKIDRHKVPTEKGA----TGLSNLGNTCFMNSSIQCVSNTQP----LTQY 761
Query 157 ISGGAATAQFAATGQQGALRRLSLALKDFYNQ-WDQTVEAVSPFLLVPTLREAYPQFSRR 215
G + T G ++ D + W T + V+P L T+ + P+F+
Sbjct 762 FISGRHLYELNRTNPIGMKGHMAKCYGDLVQELWSGTQKNVAPLKLRWTIAKYAPRFNGF 821
Query 216 STSQAGTTGVPMQQDAEECLSCLM 239
QQD++E L+ L+
Sbjct 822 Q-----------QQDSQELLAFLL 834
> mmu:30940 Usp25; ubiquitin specific peptidase 25 (EC:3.4.19.12);
K11849 ubiquitin carboxyl-terminal hydrolase 25/28 [EC:3.1.2.15]
Length=1055
Score = 37.4 bits (85), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 40/77 (51%), Gaps = 7/77 (9%)
Query 80 KIMLMGTAEEKTAVKPPPEKTVFVEDLTPAQQAALLKEKKVEPLPNGIVNLGNTCYLASV 139
+++ AE K +K P + V+ + P + K+ E P G+ N+GNTC+ ++V
Sbjct 131 RVLEASIAENKACLKRTPIE-VWRDSRNPYDR------KRQEKAPVGLKNVGNTCWFSAV 183
Query 140 LQMLRPAKDFSDLLKNY 156
+Q L +F L+ NY
Sbjct 184 IQSLFNLLEFRRLVLNY 200
> mmu:74841 Usp38, 4631402N15Rik, 4833420O05Rik, AA536967, AU044701,
AW544820, mKIAA1891; ubiquitin specific peptidase 38
(EC:3.4.19.12); K11854 ubiquitin carboxyl-terminal hydrolase
35/38 [EC:3.1.2.15]
Length=1042
Score = 37.0 bits (84), Expect = 0.056, Method: Compositional matrix adjust.
Identities = 17/31 (54%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 119 KVEPLPNGIVNLGNTCYLASVLQMLRPAKDF 149
K E G++NLGNTCY+ SVLQ L A +F
Sbjct 439 KSETGKTGLINLGNTCYMNSVLQALFMATEF 469
Lambda K H
0.316 0.130 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8402513444
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40