bitscore colors: <40, 40-50 , 50-80, 80-200, >200
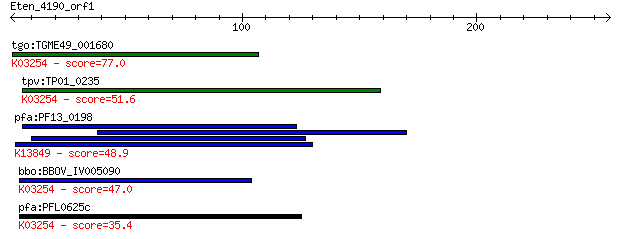
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4190_orf1
Length=256
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_001680 eukaryotic translation initiation factor 3 s... 77.0 6e-14
tpv:TP01_0235 eukaryotic translation initiation factor 3 subun... 51.6 3e-06
pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog A... 48.9 2e-05
bbo:BBOV_IV005090 23.m05782; translation initiation factor 3 s... 47.0 7e-05
pfa:PFL0625c eukaryotic translation initiation factor 3 subuni... 35.4 0.22
> tgo:TGME49_001680 eukaryotic translation initiation factor 3
subunit 10, putative (EC:3.1.2.15); K03254 translation initiation
factor 3 subunit A
Length=1033
Score = 77.0 bits (188), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 50/105 (47%), Positives = 75/105 (71%), Gaps = 0/105 (0%)
Query 2 TREDKQQWVARQLQQRQQQFAAAAAAQRQRLLQMLQLQKVQRARERMEAAIRRQRELEEQ 61
+EDK++WVA ++ RQ+ F A QR+RLL +L+ K+QRAR R E +++ + E+
Sbjct 792 VKEDKEKWVAEKIAVRQKDFEQQAQEQRERLLHVLRQAKIQRARARKEEHLKKLKIEVER 851
Query 62 RKKEEEFKRQMEEMERQKAEAEAREKRLQEIAEKQRQRELEIEAK 106
++ EEE K+ +E++E+++ E EA+ KRL E AEKQRQRELEIE K
Sbjct 852 KRMEEEQKKYLEQLEKKRQEEEAKAKRLAEQAEKQRQRELEIEEK 896
> tpv:TP01_0235 eukaryotic translation initiation factor 3 subunit
10; K03254 translation initiation factor 3 subunit A
Length=1107
Score = 51.6 bits (122), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 56/167 (33%), Positives = 84/167 (50%), Gaps = 40/167 (23%)
Query 6 KQQWVARQLQQRQQQFAAAAAAQRQRLLQMLQLQKVQRARERMEAAIRRQRELE------ 59
KQQWV +QLQ R+++ + AQ R +ML +K+ RA+ R + Q LE
Sbjct 808 KQQWVEKQLQSRKKEHQSKVDAQNARFKKMLVEEKIARAKMRYN---QEQLRLEQKRKEE 864
Query 60 ----EQRKKEEEF----KRQMEEMERQKAEAEAREKRLQEIAEKQRQRELEIEAKHGLAN 111
EQ++KEEE KR+ EE+ R++ E E +RL EIAE QRQ++LEIE
Sbjct 865 QLRLEQKRKEEELRLEQKRKEEEL-RKRREQELHRQRLDEIAEIQRQKQLEIE------- 916
Query 112 REGSRRADSQSLSHKSESPAGSSSSSSNSGQPVSPTGSRDQQQQQQQ 158
RR A +++ + NSG S + ++D + Q+
Sbjct 917 ----RRL-----------AATNNTVTVNSGYGFSNSVNKDVGRNYQK 948
> pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog
A; K13849 reticulocyte-binding protein
Length=3130
Score = 48.9 bits (115), Expect = 2e-05, Method: Composition-based stats.
Identities = 39/119 (32%), Positives = 71/119 (59%), Gaps = 10/119 (8%)
Query 6 KQQWVARQLQQRQQQFAAAAAAQRQRLLQMLQLQKVQRARERMEAAIRRQRELEEQRKKE 65
K++ + RQ Q+R Q+ A +++RL + +L++ + +ER+E RE +EQ +KE
Sbjct 2771 KEEELKRQEQERLQKEEALKRQEQERLQKEEELKR--QEQERLE------REKQEQLQKE 2822
Query 66 EEFKRQMEE-MERQKAEAEAREKRLQEIAEKQRQRELEIEAKH-GLANREGSRRADSQS 122
EE KRQ +E +++++A ++RLQ+ E +RQ + +E K LA RE ++ +S
Sbjct 2823 EELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQERLERKKIELAEREQHIKSKLES 2881
Score = 44.7 bits (104), Expect = 3e-04, Method: Composition-based stats.
Identities = 39/141 (27%), Positives = 74/141 (52%), Gaps = 23/141 (16%)
Query 38 LQKVQRARERMEAAIRRQ------RELEEQRKKEEEFKRQMEE-MERQKAEAEAREKRLQ 90
L++ ++ R + E ++RQ RE +EQ +KEEE KRQ +E +++++A ++RLQ
Sbjct 2739 LKRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQ 2798
Query 91 EIAE--KQRQRELEIEAKHGLANREGSRRADSQSLSHKSESPAGSSSSSSNSGQPVSPTG 148
+ E +Q Q LE E + L E +R + + L K E+
Sbjct 2799 KEEELKRQEQERLEREKQEQLQKEEELKRQEQERL-QKEEALKRQEQ------------- 2844
Query 149 SRDQQQQQQQQQQQQQLEWRR 169
R Q++++ ++Q+Q++LE ++
Sbjct 2845 ERLQKEEELKRQEQERLERKK 2865
Score = 44.3 bits (103), Expect = 5e-04, Method: Composition-based stats.
Identities = 38/126 (30%), Positives = 68/126 (53%), Gaps = 9/126 (7%)
Query 10 VARQLQQRQQQFAAAAAAQRQRL--LQMLQLQKVQ----RARERMEAAIRRQRELEEQRK 63
+ RQ Q+R Q+ +++RL + QLQK + + +ER++ +R+ +E+ +
Sbjct 2739 LKRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQ 2798
Query 64 KEEEFKRQMEE-MERQKAEAEAREKRL--QEIAEKQRQRELEIEAKHGLANREGSRRADS 120
KEEE KRQ +E +ER+K E +E+ L QE Q++ L+ + + L E +R +
Sbjct 2799 KEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQ 2858
Query 121 QSLSHK 126
+ L K
Sbjct 2859 ERLERK 2864
Score = 43.9 bits (102), Expect = 5e-04, Method: Composition-based stats.
Identities = 31/129 (24%), Positives = 71/129 (55%), Gaps = 3/129 (2%)
Query 3 REDKQQWVARQLQQRQQQFAAAAAAQRQRLLQMLQLQKVQRARERMEAAIRRQRELEEQR 62
+ +Q+ + R+ Q++ Q+ +++RL + L++ ++ R + E ++RQ + +R
Sbjct 2754 KRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQERLER 2813
Query 63 KKEEEFKRQMEEMERQKAEAEAREKRL--QEIAEKQRQRELEIEAKHGLANREGSRRADS 120
+K+E+ +++ EE++RQ+ E +E+ L QE Q++ EL+ + + L ++
Sbjct 2814 EKQEQLQKE-EELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQERLERKKIELAERE 2872
Query 121 QSLSHKSES 129
Q + K ES
Sbjct 2873 QHIKSKLES 2881
> bbo:BBOV_IV005090 23.m05782; translation initiation factor 3
subunit 10; K03254 translation initiation factor 3 subunit
A
Length=991
Score = 47.0 bits (110), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 44/99 (44%), Positives = 68/99 (68%), Gaps = 0/99 (0%)
Query 5 DKQQWVARQLQQRQQQFAAAAAAQRQRLLQMLQLQKVQRARERMEAAIRRQRELEEQRKK 64
+K++W+ + +RQ + A QR+R ++L K+QRARER + +RRQ+ELEE+R +
Sbjct 804 EKEKWINDTMARRQAIYDAEIEEQRKRFTEILIKDKIQRARERRDYELRRQKELEEERIR 863
Query 65 EEEFKRQMEEMERQKAEAEAREKRLQEIAEKQRQRELEI 103
EEE R+ E +R++ E R+++L EIAEKQR +ELEI
Sbjct 864 EEERMRREMEEKRKQEEEAERKRKLMEIAEKQRAKELEI 902
> pfa:PFL0625c eukaryotic translation initiation factor 3 subunit
10, putative; K03254 translation initiation factor 3 subunit
A
Length=1377
Score = 35.4 bits (80), Expect = 0.22, Method: Composition-based stats.
Identities = 36/120 (30%), Positives = 74/120 (61%), Gaps = 1/120 (0%)
Query 5 DKQQWVARQLQQRQQQFAAAAAAQRQRLLQMLQLQKVQRARERMEAAIRRQRELEEQRKK 64
D +++ +++ R ++F AQ++RL +L+ +K+QRA+ER + IR+ + EE++++
Sbjct 910 DIEEFTKNEMKLRVEEFEKNLKAQKERLYNILKEEKIQRAKERRDQHIRKLKAEEERKRR 969
Query 65 EEEFKRQMEEMERQKAEAEAREKRLQEIAEKQRQRELEIEAKHGLANREGSRRADSQSLS 124
EEE + ++ + E ++ + E K+L+EI++ QR+R+LE++ + E R S S
Sbjct 970 EEE-EERLLKEEMERKKKEEARKKLEEISKAQRERDLEMDLQMKRKKEESRRSERSHRKS 1028
Lambda K H
0.308 0.118 0.313
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9340616888
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40