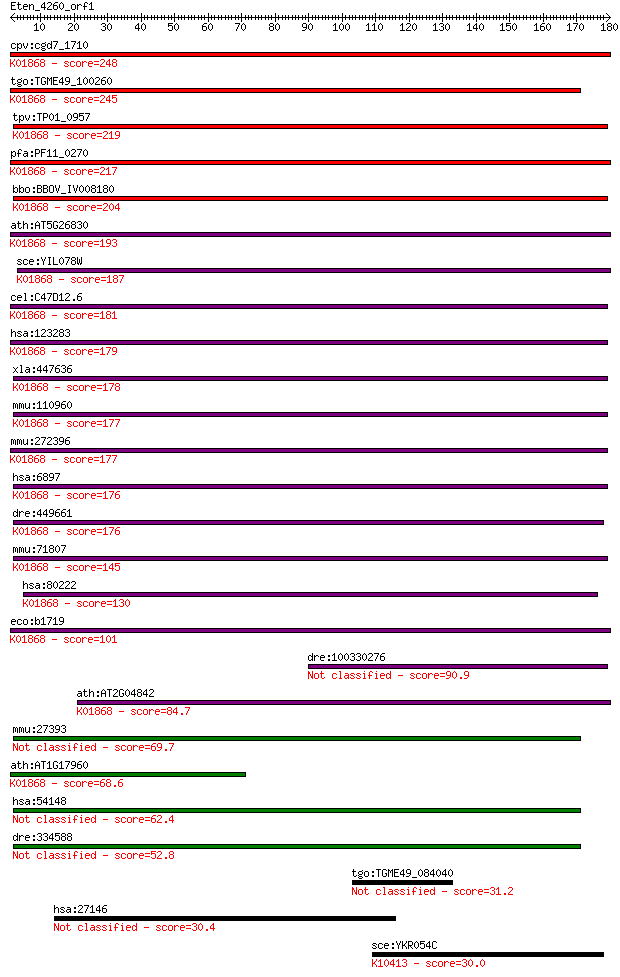
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4260_orf1
Length=179
Score E
Sequences producing significant alignments: (Bits) Value
cpv:cgd7_1710 threonyl-tRNA synthetase (RNA binding domain TGS... 248 7e-66
tgo:TGME49_100260 threonyl-tRNA synthetase, putative (EC:6.1.1... 245 6e-65
tpv:TP01_0957 threonyl-tRNA synthetase; K01868 threonyl-tRNA s... 219 3e-57
pfa:PF11_0270 threonine --tRNA ligase, putative; K01868 threon... 217 1e-56
bbo:BBOV_IV008180 23.m05860; threonyl-tRNA synthetase (EC:6.1.... 204 1e-52
ath:AT5G26830 threonyl-tRNA synthetase / threonine--tRNA ligas... 193 2e-49
sce:YIL078W THS1; Ths1p (EC:6.1.1.3); K01868 threonyl-tRNA syn... 187 2e-47
cel:C47D12.6 trs-1; Threonyl tRNA Synthetase family member (tr... 181 1e-45
hsa:123283 TARSL2, FLJ25005; threonyl-tRNA synthetase-like 2 (... 179 5e-45
xla:447636 tars, MGC86352; threonyl-tRNA synthetase (EC:6.1.1.... 178 8e-45
mmu:110960 Tars, D15Wsu59e, ThrRS; threonyl-tRNA synthetase (E... 177 2e-44
mmu:272396 Tarsl2, A530046H20Rik, MGC31414; threonyl-tRNA synt... 177 3e-44
hsa:6897 TARS, MGC9344, ThrRS; threonyl-tRNA synthetase (EC:6.... 176 3e-44
dre:449661 tars, ThrRS, wu:fb07c05, wu:fb39c12, zgc:92586; thr... 176 3e-44
mmu:71807 Tars2, 2610024N01Rik, AI429208, Tarsl1; threonyl-tRN... 145 8e-35
hsa:80222 TARS2, FLJ12528, TARSL1; threonyl-tRNA synthetase 2,... 130 3e-30
eco:b1719 thrS, ECK1717, JW1709; threonyl-tRNA synthetase (EC:... 101 2e-21
dre:100330276 threonyl-tRNA synthetase-like 90.9 2e-18
ath:AT2G04842 EMB2761 (EMBRYO DEFECTIVE 2761); ATP binding / a... 84.7 2e-16
mmu:27393 Mrpl39, C21orf8, MRP-L5, ORF22, Rpml5; mitochondrial... 69.7 5e-12
ath:AT1G17960 threonyl-tRNA synthetase, putative / threonine--... 68.6 1e-11
hsa:54148 MRPL39, C21orf92, FLJ20451, L39mt, MGC104174, MGC340... 62.4 8e-10
dre:334588 mrpl39, fa13h09, wu:fa13h09, zgc:63888; mitochondri... 52.8 6e-07
tgo:TGME49_084040 hypothetical protein 31.2 1.7
hsa:27146 FAM184B, KIAA1276; family with sequence similarity 1... 30.4 3.3
sce:YKR054C DYN1, DHC1, PAC6; Cytoplasmic heavy chain dynein, ... 30.0 4.5
> cpv:cgd7_1710 threonyl-tRNA synthetase (RNA binding domain TGS+HxxxH+tRNA
synthetase) ; K01868 threonyl-tRNA synthetase
[EC:6.1.1.3]
Length=763
Score = 248 bits (633), Expect = 7e-66, Method: Compositional matrix adjust.
Identities = 109/179 (60%), Positives = 138/179 (77%), Gaps = 0/179 (0%)
Query 1 GSCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSL 60
G C+L+IL F S E VFWHSS+HILGQ +E E+GA +T+GPAL PGFYYD YMG S+
Sbjct 146 GDCELEILTFDSLEGSGVFWHSSSHILGQCLENEYGAQVTIGPALNPGFYYDSYMGTHSV 205
Query 61 SESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYR 120
S +E+ + A+ I+SE Q FERL C KEEALE+F +NPFKV+LI +K+PDG TT+YR
Sbjct 206 SNTEYSDIENCAKTIISEKQQFERLQCNKEEALELFKDNPFKVSLIMSKIPDGAQTTIYR 265
Query 121 CGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEYL 179
CG+ VD+C GPH+P T + K+F VT++S WLGN ND+LQRVYGVSFP+KK+L EYL
Sbjct 266 CGSFVDLCTGPHIPHTGIVKAFKVTKNSGCNWLGNTENDALQRVYGVSFPDKKRLDEYL 324
> tgo:TGME49_100260 threonyl-tRNA synthetase, putative (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=844
Score = 245 bits (625), Expect = 6e-65, Method: Compositional matrix adjust.
Identities = 112/170 (65%), Positives = 136/170 (80%), Gaps = 0/170 (0%)
Query 1 GSCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSL 60
GSC+LQ+LKF S EA++VFWHSSAHILGQA+E FGA LTVGPAL GFYYD YMG +
Sbjct 208 GSCRLQLLKFDSDEAKHVFWHSSAHILGQAIEATFGAQLTVGPALTNGFYYDAYMGDAKV 267
Query 61 SESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYR 120
+E +G L AAA ++ E Q F RLVC+K EALE+FA+NPFKV LI +K+P+ GLTTVY
Sbjct 268 TEESYGRLEAAAAAVIKEDQAFRRLVCSKAEALELFADNPFKVQLIASKIPEHGLTTVYC 327
Query 121 CGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFP 170
CG+LVD+CRGPH+P+T K+F V +HSA+YWLG + DSLQRVYGVSFP
Sbjct 328 CGSLVDLCRGPHIPSTGKVKAFQVIKHSASYWLGRQHLDSLQRVYGVSFP 377
> tpv:TP01_0957 threonyl-tRNA synthetase; K01868 threonyl-tRNA
synthetase [EC:6.1.1.3]
Length=779
Score = 219 bits (559), Expect = 3e-57, Method: Compositional matrix adjust.
Identities = 101/179 (56%), Positives = 128/179 (71%), Gaps = 2/179 (1%)
Query 2 SCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSLS 61
SCK++ L F S + Q+V+WHSSAHILG A+E FG LT+GPAL GFYYD Y+G S+
Sbjct 189 SCKVEFLDFNSEQGQHVYWHSSAHILGSALETCFGGFLTIGPALSSGFYYDVYLGNNSVK 248
Query 62 ESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYRC 121
+ L + + + + PFERLVCTKEEALE+ NPFKV LI KVPDG T YRC
Sbjct 249 PEDVKPLLSHVEALTNLNSPFERLVCTKEEALELMKYNPFKVQLIKNKVPDGENTVCYRC 308
Query 122 GTLVDICRGPHVPTTALCK--SFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEY 178
G VD+CRGPH+PTT++ + SF VT+ S++YWL N +D+LQRVYG+SFPEK+QL Y
Sbjct 309 GDFVDLCRGPHIPTTSMVRANSFDVTKISSSYWLSNAKSDTLQRVYGISFPEKEQLKMY 367
> pfa:PF11_0270 threonine --tRNA ligase, putative; K01868 threonyl-tRNA
synthetase [EC:6.1.1.3]
Length=1013
Score = 217 bits (553), Expect = 1e-56, Method: Composition-based stats.
Identities = 90/179 (50%), Positives = 128/179 (71%), Gaps = 0/179 (0%)
Query 1 GSCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSL 60
G+C+++ + S E Q +FWHSSAHILG ++E FG LT+GPALK GFYYD ++ S+
Sbjct 342 GNCRVEFINIQSEEGQKIFWHSSAHILGSSLEKLFGGFLTIGPALKEGFYYDIFLNNFSI 401
Query 61 SESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYR 120
+ ++ + ++V + PFE+++CTKEEALE+F NPFK+ LI +K+PD T+VYR
Sbjct 402 NNEDYKRIEDEFNKLVKDNVPFEKVICTKEEALELFDYNPFKLELIRSKIPDNKKTSVYR 461
Query 121 CGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEYL 179
CG +D+C GPH+ T K+F V ++S+AYWLG + NDSLQRVYG+SF +K +L EYL
Sbjct 462 CGNFIDLCLGPHIKNTGKVKTFKVLKNSSAYWLGQKENDSLQRVYGISFQKKSELVEYL 520
> bbo:BBOV_IV008180 23.m05860; threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=736
Score = 204 bits (520), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 91/179 (50%), Positives = 119/179 (66%), Gaps = 2/179 (1%)
Query 2 SCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSLS 61
C ++ F SP+ Q+VFWHSSAH+LG A+E +GA +T+GPAL GFYYDCY+G +
Sbjct 136 DCTVRFHTFESPKGQHVFWHSSAHLLGSALETLYGAYITIGPALSQGFYYDCYLGNNTFK 195
Query 62 ESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYRC 121
+ + + PF+RLVC+KEEALE+ NPFKV LI KVPDG T YRC
Sbjct 196 PDDMALITEQVSTVTKTKSPFQRLVCSKEEALELMKHNPFKVELIRKKVPDGSSTVCYRC 255
Query 122 GTLVDICRGPHVP--TTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEY 178
G VD+CRGPH+P T+ K+ +VT+ S+ YWL + DSLQRVYG+SFP++ QL Y
Sbjct 256 GDFVDLCRGPHLPFTTSVHHKAMTVTKFSSCYWLSKNTADSLQRVYGISFPKEAQLKAY 314
> ath:AT5G26830 threonyl-tRNA synthetase / threonine--tRNA ligase
(THRRS) (EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=709
Score = 193 bits (491), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 94/181 (51%), Positives = 127/181 (70%), Gaps = 3/181 (1%)
Query 1 GSCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALK--PGFYYDCYMGQK 58
G CKL++ KF S + ++ WHSSAHILGQA+E E+G L +GP GFYYD + G+
Sbjct 127 GDCKLELFKFDSDKGRDTLWHSSAHILGQALEQEYGCQLCIGPCTTRGEGFYYDGFYGEL 186
Query 59 SLSESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTV 118
LS++ ++ A A + EAQPFER+ TK++ALEMF+EN FKV LI +P TV
Sbjct 187 GLSDNHFPSIEAGAAKAAKEAQPFERIEVTKDQALEMFSENNFKVELING-LPADMTITV 245
Query 119 YRCGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEY 178
YRCG LVD+CRGPH+P T+ K+F R S+AYW G++ +SLQRVYG+S+P++KQL +Y
Sbjct 246 YRCGPLVDLCRGPHIPNTSFVKAFKCLRASSAYWKGDKDRESLQRVYGISYPDQKQLKKY 305
Query 179 L 179
L
Sbjct 306 L 306
> sce:YIL078W THS1; Ths1p (EC:6.1.1.3); K01868 threonyl-tRNA synthetase
[EC:6.1.1.3]
Length=734
Score = 187 bits (475), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 84/187 (44%), Positives = 127/187 (67%), Gaps = 10/187 (5%)
Query 3 CKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMG------ 56
KL++L F S E + VFWHSSAH+LG++ EC GA + +GP GF+Y+ +
Sbjct 133 IKLELLDFESDEGKKVFWHSSAHVLGESCECHLGAHICLGPPTDDGFFYEMAVRDSMKDI 192
Query 57 ----QKSLSESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPD 112
++++S+++ L A+ ++ + Q FERLV +KE+ L+MF + +K L+ TKVPD
Sbjct 193 SESPERTVSQADFPGLEGVAKNVIKQKQKFERLVMSKEDLLKMFHYSKYKTYLVQTKVPD 252
Query 113 GGLTTVYRCGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEK 172
GG TTVYRCG L+D+C GPH+P T K+F + ++S+ Y+LG+ +NDSLQRVYG+SFP+K
Sbjct 253 GGATTVYRCGKLIDLCVGPHIPHTGRIKAFKLLKNSSCYFLGDATNDSLQRVYGISFPDK 312
Query 173 KQLTEYL 179
K + +L
Sbjct 313 KLMDAHL 319
> cel:C47D12.6 trs-1; Threonyl tRNA Synthetase family member (trs-1);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=725
Score = 181 bits (458), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 85/178 (47%), Positives = 119/178 (66%), Gaps = 1/178 (0%)
Query 1 GSCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSL 60
G+ KL++LKF EA+ VFWHSSAH+LG+AME G L GP ++ GFYYD + +++
Sbjct 134 GNAKLELLKFDDDEAKQVFWHSSAHVLGEAMERYCGGHLCYGPPIQEGFYYDMWHENRTI 193
Query 61 SESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYR 120
+ ++ + V + Q FERL TKE+ LEMF N FKV +IT K+ TTVYR
Sbjct 194 CPDDFPKIDQIVKAAVKDKQKFERLEMTKEDLLEMFKYNEFKVRIITEKIHTPK-TTVYR 252
Query 121 CGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEY 178
CG L+D+CRGPHV T K+ ++T++S++YW G +SLQR+YG+SFP+ KQL E+
Sbjct 253 CGPLIDLCRGPHVRHTGKVKAMAITKNSSSYWEGKADAESLQRLYGISFPDSKQLKEW 310
> hsa:123283 TARSL2, FLJ25005; threonyl-tRNA synthetase-like 2
(EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=802
Score = 179 bits (454), Expect = 5e-45, Method: Compositional matrix adjust.
Identities = 82/178 (46%), Positives = 118/178 (66%), Gaps = 1/178 (0%)
Query 1 GSCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSL 60
G L++L F + EAQ V+WHSSAHILG+AME +G L GP ++ GFYYD ++ +++
Sbjct 214 GDSSLELLTFDNEEAQAVYWHSSAHILGEAMELYYGGHLCYGPPIENGFYYDMFIEDRAV 273
Query 61 SESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYR 120
S +E AL + I+ E QPFERL +KE LEMF N FK ++ KV + TTVYR
Sbjct 274 SSTELSALENICKAIIKEKQPFERLEVSKEILLEMFKYNKFKCRILNEKV-NTATTTVYR 332
Query 121 CGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEY 178
CG L+D+C+GPHV T K+ + ++S+ YW GN ++LQR+YG+SFP+ K + ++
Sbjct 333 CGPLIDLCKGPHVRHTGKIKTIKIFKNSSTYWEGNPEMETLQRIYGISFPDNKMMRDW 390
> xla:447636 tars, MGC86352; threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=721
Score = 178 bits (452), Expect = 8e-45, Method: Compositional matrix adjust.
Identities = 85/177 (48%), Positives = 114/177 (64%), Gaps = 1/177 (0%)
Query 2 SCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSLS 61
C L +LKF EAQ V+WHSSAHI+G+AME +G L GP ++ GFYYD Y+ +S
Sbjct 134 DCTLALLKFDDEEAQAVYWHSSAHIMGEAMERVYGGCLCYGPPIENGFYYDMYLDDGGVS 193
Query 62 ESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYRC 121
++ +L ++I+ E Q FERL +KE LEMF N FK ++ KV D TTVYRC
Sbjct 194 SNDFTSLENLCKKIMKEKQQFERLEVSKETLLEMFKYNKFKCRILNEKV-DTPTTTVYRC 252
Query 122 GTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEY 178
G L+D+CRGPHV T K+ V ++S+ YW G ++LQRVYG+SFP+ K L E+
Sbjct 253 GPLIDLCRGPHVRHTGKIKALKVHKNSSTYWEGKADMETLQRVYGISFPDPKMLKEW 309
> mmu:110960 Tars, D15Wsu59e, ThrRS; threonyl-tRNA synthetase
(EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=722
Score = 177 bits (448), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 82/177 (46%), Positives = 115/177 (64%), Gaps = 1/177 (0%)
Query 2 SCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSLS 61
C L++LKF EAQ V+WHSSAHI+G+AME +G L GP ++ GFYYD Y+ + +S
Sbjct 135 DCTLELLKFEDEEAQAVYWHSSAHIMGEAMERVYGGCLCYGPPIENGFYYDMYLEEGGVS 194
Query 62 ESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYRC 121
++ +L ++I+ E Q FERL KE LEMF N FK ++ KV + TTVYRC
Sbjct 195 SNDFSSLETLCKKIIKEKQTFERLEVKKETLLEMFKYNKFKCRILNEKV-NTPTTTVYRC 253
Query 122 GTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEY 178
G L+D+CRGPHV T K+ + ++S+ YW G ++LQR+YG+SFP+ K L E+
Sbjct 254 GPLIDLCRGPHVRHTGKIKTLKIHKNSSTYWEGKADMETLQRIYGISFPDPKLLKEW 310
> mmu:272396 Tarsl2, A530046H20Rik, MGC31414; threonyl-tRNA synthetase-like
2 (EC:6.1.1.3); K01868 threonyl-tRNA synthetase
[EC:6.1.1.3]
Length=790
Score = 177 bits (448), Expect = 3e-44, Method: Compositional matrix adjust.
Identities = 81/178 (45%), Positives = 117/178 (65%), Gaps = 1/178 (0%)
Query 1 GSCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSL 60
G +++L F + EAQ V+WHSSAHILG+AME +G L GP ++ GFYYD ++ + +
Sbjct 202 GDSTVELLMFDNEEAQAVYWHSSAHILGEAMELYYGGHLCYGPPIENGFYYDMFIEDRVV 261
Query 61 SESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYR 120
S +E AL + I+ E QPFERL +K+ LEMF N FK ++ KV D TTVYR
Sbjct 262 SSTELSALENICKTIIKEKQPFERLEVSKDTLLEMFKYNKFKCRILKEKV-DTPTTTVYR 320
Query 121 CGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEY 178
CG L+D+C+GPHV T K+ + ++S+ YW GN ++LQR+YG+SFP+ K + ++
Sbjct 321 CGPLIDLCKGPHVRHTGKIKAIKIFKNSSTYWEGNPEMETLQRIYGISFPDSKMMKDW 378
> hsa:6897 TARS, MGC9344, ThrRS; threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=723
Score = 176 bits (447), Expect = 3e-44, Method: Compositional matrix adjust.
Identities = 82/177 (46%), Positives = 115/177 (64%), Gaps = 1/177 (0%)
Query 2 SCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSLS 61
C L++LKF EAQ V+WHSSAHI+G+AME +G L GP ++ GFYYD Y+ + +S
Sbjct 136 DCTLELLKFEDEEAQAVYWHSSAHIMGEAMERVYGGCLCYGPPIENGFYYDMYLEEGGVS 195
Query 62 ESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYRC 121
++ +L A ++I+ E Q FERL KE L MF N FK ++ KV + TTVYRC
Sbjct 196 SNDFSSLEALCKKIIKEKQAFERLEVKKETLLAMFKYNKFKCRILNEKV-NTPTTTVYRC 254
Query 122 GTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEY 178
G L+D+CRGPHV T K+ + ++S+ YW G ++LQR+YG+SFP+ K L E+
Sbjct 255 GPLIDLCRGPHVRHTGKIKALKIHKNSSTYWEGKADMETLQRIYGISFPDPKMLKEW 311
> dre:449661 tars, ThrRS, wu:fb07c05, wu:fb39c12, zgc:92586; threonyl-tRNA
synthetase (EC:6.1.1.3); K01868 threonyl-tRNA synthetase
[EC:6.1.1.3]
Length=624
Score = 176 bits (447), Expect = 3e-44, Method: Compositional matrix adjust.
Identities = 84/176 (47%), Positives = 111/176 (63%), Gaps = 1/176 (0%)
Query 2 SCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSLS 61
C L LKF EAQ V+WHSSAHI+G+AME +G L GP ++ GFYYD Y+ + +S
Sbjct 131 DCSLVFLKFDDEEAQAVYWHSSAHIMGEAMERVYGGCLCYGPPIENGFYYDMYLDNEGVS 190
Query 62 ESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYRC 121
++ L ++I+ E QPFERL KE LEMF N FK ++ KV TTVYRC
Sbjct 191 SNDFPCLENLCKKIIKEKQPFERLEVKKETLLEMFKYNKFKCRILNEKVT-TPTTTVYRC 249
Query 122 GTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTE 177
G L+D+CRGPHV T K+ + ++S+ YW G +SLQR+YG+SFP+ K L E
Sbjct 250 GPLIDLCRGPHVRHTGKIKAMKIHKNSSTYWEGKADMESLQRIYGISFPDPKMLKE 305
> mmu:71807 Tars2, 2610024N01Rik, AI429208, Tarsl1; threonyl-tRNA
synthetase 2, mitochondrial (putative) (EC:6.1.1.3); K01868
threonyl-tRNA synthetase [EC:6.1.1.3]
Length=697
Score = 145 bits (365), Expect = 8e-35, Method: Compositional matrix adjust.
Identities = 72/178 (40%), Positives = 111/178 (62%), Gaps = 2/178 (1%)
Query 2 SCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMG-QKSL 60
C L+ L F SPE + VFWHSSAH+LG A E + GA+L GP+ + GFY+D ++G ++++
Sbjct 119 DCHLRFLTFDSPEGKAVFWHSSAHVLGAAAEQQLGAVLCRGPSTESGFYHDFFLGKERTV 178
Query 61 SESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYR 120
+E L Q +++ AQPF RL ++++ ++F +N FK+ LI KV G TVY
Sbjct 179 RSAELPILERICQELIAAAQPFRRLEASRDQLRQLFKDNHFKLHLIEEKV-TGPTATVYG 237
Query 121 CGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEY 178
CG VD+CRGPH+ T + + +S+A W + ++LQRV G+SFP+ + L +
Sbjct 238 CGMSVDLCRGPHLRHTGQIGALKLLTNSSALWRSLGAPETLQRVSGISFPKVELLRNW 295
> hsa:80222 TARS2, FLJ12528, TARSL1; threonyl-tRNA synthetase
2, mitochondrial (putative) (EC:6.1.1.3); K01868 threonyl-tRNA
synthetase [EC:6.1.1.3]
Length=718
Score = 130 bits (326), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 71/172 (41%), Positives = 108/172 (62%), Gaps = 2/172 (1%)
Query 5 LQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMG-QKSLSES 63
L+ L F SPE + VFWHSS H+LG A E GA+L GP+ + GFY+D ++G ++++ S
Sbjct 117 LRFLTFDSPEGKAVFWHSSTHVLGAAAEQFLGAVLCRGPSTEYGFYHDFFLGKERTIRGS 176
Query 64 EHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYRCGT 123
E L Q + + A+PF RL ++++ ++F +NPFK+ LI KV G TVY CGT
Sbjct 177 ELPVLERICQELTAAARPFRRLEASRDQLRQLFKDNPFKLHLIEEKV-TGPTATVYGCGT 235
Query 124 LVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQL 175
LVD+C+GPH+ T + +S++ W + + ++LQRV G+SFP + L
Sbjct 236 LVDLCQGPHLRHTGQIGGLKLLSNSSSLWRSSGAPETLQRVSGISFPTTELL 287
> eco:b1719 thrS, ECK1717, JW1709; threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=642
Score = 101 bits (251), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 54/182 (29%), Positives = 90/182 (49%), Gaps = 4/182 (2%)
Query 1 GSCKLQILKFVSPEAQNVFWHSSAHILGQAMECEF-GALLTVGPALKPGFYYDCYMGQKS 59
+L I+ E + HS AH+LG A++ + + +GP + GFYYD + ++
Sbjct 53 NDAQLSIITAKDEEGLEIIRHSCAHLLGHAIKQLWPHTKMAIGPVIDNGFYYDVDL-DRT 111
Query 60 LSESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAEN--PFKVALITTKVPDGGLTT 117
L++ + AL + + + + EA E FA +KV+++ +
Sbjct 112 LTQEDVEALEKRMHELAEKNYDVIKKKVSWHEARETFANRGESYKVSILDENIAHDDKPG 171
Query 118 VYRCGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTE 177
+Y VD+CRGPHVP C F + + + AYW G+ +N LQR+YG ++ +KK L
Sbjct 172 LYFHEEYVDMCRGPHVPNMRFCHHFKLMKTAGAYWRGDSNNKMLQRIYGTAWADKKALNA 231
Query 178 YL 179
YL
Sbjct 232 YL 233
> dre:100330276 threonyl-tRNA synthetase-like
Length=380
Score = 90.9 bits (224), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 41/89 (46%), Positives = 57/89 (64%), Gaps = 1/89 (1%)
Query 90 EEALEMFAENPFKVALITTKVPDGGLTTVYRCGTLVDICRGPHVPTTALCKSFSVTRHSA 149
+E L F N FK ++ KV TTVYRCG L+D+CRGPHV T K+F + ++S+
Sbjct 24 KEQLFFFQYNKFKCRILNEKVT-TDTTTVYRCGPLIDLCRGPHVRHTGKIKAFKIYKNSS 82
Query 150 AYWLGNRSNDSLQRVYGVSFPEKKQLTEY 178
YW G ++LQR+YG+SFP+ K L E+
Sbjct 83 TYWEGRSDMETLQRIYGISFPDSKMLKEW 111
> ath:AT2G04842 EMB2761 (EMBRYO DEFECTIVE 2761); ATP binding /
aminoacyl-tRNA ligase/ ligase, forming aminoacyl-tRNA and related
compounds / nucleotide binding / threonine-tRNA ligase
(EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=650
Score = 84.7 bits (208), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 54/165 (32%), Positives = 84/165 (50%), Gaps = 10/165 (6%)
Query 21 HSSAHILGQAMECEF-GALLTVGPALKPGFYYDCYMGQKSLSESEHGALNAAAQRIVSEA 79
H+ AH++ A++ F A +T+GP + GFYYD M + L++ + + RI+S
Sbjct 84 HTCAHVMAMAVQKLFPDAKVTIGPWIDNGFYYDFDM--EPLTDKDLKRIKKEMDRIISRN 141
Query 80 QPFERLVCTKEEALE--MFAENPFKVALITTKVPDGGLTTVYRCGT-LVDICRGPHVPTT 136
P R ++EEA + M P+K+ ++ + TVY G D+C GPHV TT
Sbjct 142 LPLLREEVSREEAKKRIMAINEPYKMEILDGIKEEP--ITVYHIGNEWWDLCAGPHVETT 199
Query 137 ALCKSFSVTRHS--AAYWLGNRSNDSLQRVYGVSFPEKKQLTEYL 179
+V S AYW G+ LQR+YG ++ ++QL YL
Sbjct 200 GKINKKAVELESVAGAYWRGDEKRQMLQRIYGTAWESEEQLKAYL 244
> mmu:27393 Mrpl39, C21orf8, MRP-L5, ORF22, Rpml5; mitochondrial
ribosomal protein L39
Length=297
Score = 69.7 bits (169), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 48/185 (25%), Positives = 81/185 (43%), Gaps = 19/185 (10%)
Query 2 SCKLQILKFVSPEAQNV---FWHSSAHILGQAMECEFGALLTVG-------PALKPGFYY 51
SC+++ L F P+ + V +W S A +LG +E F V P + F Y
Sbjct 80 SCEIKFLTFKDPDPKEVNKAYWRSCAMMLGCVIERAFKDDYVVSLVRAPEVPVIAGAFCY 139
Query 52 DCYMGQK----SLSESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALIT 107
D + ++ ++ + A ++ PFE L ALE+F N +KV I
Sbjct 140 DVTLDKRLDEWMPTKENLRSFTKDAHALIYRDLPFETLDVDARVALEIFQHNKYKVDFIE 199
Query 108 TKVPDG--GLTTVYRCGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVY 165
K + ++R G +D+ GP +P T++C + V SA + L + ++R
Sbjct 200 EKASQNPERIVKLHRIGDFIDVSEGPLIPRTSVCFQYEV---SAVHNLNPSQPNLIRRFQ 256
Query 166 GVSFP 170
G+S P
Sbjct 257 GLSLP 261
> ath:AT1G17960 threonyl-tRNA synthetase, putative / threonine--tRNA
ligase, putative; K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=458
Score = 68.6 bits (166), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 34/73 (46%), Positives = 45/73 (61%), Gaps = 3/73 (4%)
Query 1 GSCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGP--ALKPGFYYDC-YMGQ 57
G C L+I F S + +N FWHSSAHILGQA+E E+G L +GP GFYYD Y G+
Sbjct 96 GDCSLEIFGFDSDQGRNTFWHSSAHILGQALEQEYGCKLCIGPCEPRDEGFYYDSLYYGE 155
Query 58 KSLSESEHGALNA 70
L+++ + A
Sbjct 156 LGLNDNHFPNIEA 168
> hsa:54148 MRPL39, C21orf92, FLJ20451, L39mt, MGC104174, MGC3400,
MRP-L5, MRPL5, PRED22, PRED66, RPML5; mitochondrial ribosomal
protein L39
Length=338
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 48/185 (25%), Positives = 79/185 (42%), Gaps = 19/185 (10%)
Query 2 SCKLQILKF--VSP-EAQNVFWHSSAHILGQAMECEFGALLTVG-------PALKPGFYY 51
SC+++ L F P E +W S A ++G +E F V P + F Y
Sbjct 122 SCEIKFLTFKDCDPGEVNKAYWRSCAMMMGCVIERAFKDEYMVNLVRAPEVPVISGAFCY 181
Query 52 DCYMGQK----SLSESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALIT 107
D + K ++ + A ++ + PFE L + ALE+F + +KV I
Sbjct 182 DVVLDSKLDEWMPTKENLRSFTKDAHALIYKDLPFETLEVEAKVALEIFQHSKYKVDFIE 241
Query 108 TKVPDG--GLTTVYRCGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVY 165
K + ++R G +D+ GP +P T++C + V SA + L ++R
Sbjct 242 EKASQNPERIVKLHRIGDFIDVSEGPLIPRTSICFQYEV---SAVHNLQPTQPSLIRRFQ 298
Query 166 GVSFP 170
GVS P
Sbjct 299 GVSLP 303
> dre:334588 mrpl39, fa13h09, wu:fa13h09, zgc:63888; mitochondrial
ribosomal protein L39
Length=342
Score = 52.8 bits (125), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 49/185 (26%), Positives = 78/185 (42%), Gaps = 19/185 (10%)
Query 2 SCKLQILKFVSPEAQNV---FWHSSAHILGQAMECEFGALLTVGPALKPGF-----YYDC 53
SC+L +L F + Q + +W S A +LG +E F +V P + C
Sbjct 112 SCELTLLTFRDADPQTLNQAYWRSCAALLGLVLESAFKDQFSVELLKTPEVPVTAGAFCC 171
Query 54 YMGQKSL------SESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALIT 107
+ L +E +L A +++S P+E L + ALE+F+++ K +
Sbjct 172 DLLLDPLLDSWKPTEENLRSLTRDALQMISRDLPWEPLEVSASLALEIFSQSRCKQEEVE 231
Query 108 TKV--PDGGLTTVYRCGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVY 165
G T+YRCG V + P V T LC F VT + + L + +R
Sbjct 232 EAAAQSQNGTVTLYRCGDHVTLSSAPLVSRTGLCSQFEVT---SIHTLSEHTRGVQRRAQ 288
Query 166 GVSFP 170
G+S P
Sbjct 289 GLSLP 293
> tgo:TGME49_084040 hypothetical protein
Length=1841
Score = 31.2 bits (69), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 17/32 (53%), Gaps = 2/32 (6%)
Query 103 VALITTKVPDGGLT--TVYRCGTLVDICRGPH 132
AL + P GGL+ TV C VD CR PH
Sbjct 938 AALSRSPPPRGGLSPSTVDACARFVDACRAPH 969
> hsa:27146 FAM184B, KIAA1276; family with sequence similarity
184, member B
Length=1060
Score = 30.4 bits (67), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 27/104 (25%), Positives = 47/104 (45%), Gaps = 2/104 (1%)
Query 14 EAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSLSESEHGALNAAAQ 73
E QN + A +L + E ALL AL+ + +++L+ES L +
Sbjct 73 ELQNAVAETKARLLQEQGCAEEEALLQRIQALESALELQKRLTEEALAESASCRLETKER 132
Query 74 RIVSEAQPFERLVCTKEEALEMFAENPFKVALITTK--VPDGGL 115
+ EA+ ER++ E LE+ A+ ++ +T+ P G L
Sbjct 133 ELRVEAEHAERVLTLSREMLELKADYERRLQHLTSHEATPQGRL 176
> sce:YKR054C DYN1, DHC1, PAC6; Cytoplasmic heavy chain dynein,
microtubule motor protein, required for anaphase spindle elongation;
involved in spindle assembly, chromosome movement,
and spindle orientation during cell division, targeted to
microtubule tips by Pac1p; K10413 dynein heavy chain 1, cytosolic
Length=4092
Score = 30.0 bits (66), Expect = 4.5, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 34/76 (44%), Gaps = 9/76 (11%)
Query 109 KVPDGGLTTVYRCGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVS 168
K P+ T+ R +V C P P + S TRH+A +LG S SL ++Y +
Sbjct 2517 KTPENKWVTIERI-HIVGACNPPTDPGR-IPMSERFTRHAAILYLGYPSGKSLSQIYEIY 2574
Query 169 F-------PEKKQLTE 177
+ PE + TE
Sbjct 2575 YKAIFKLVPEFRSYTE 2590
Lambda K H
0.320 0.134 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4795148792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40