bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4284_orf3
Length=187
Score E
Sequences producing significant alignments: (Bits) Value
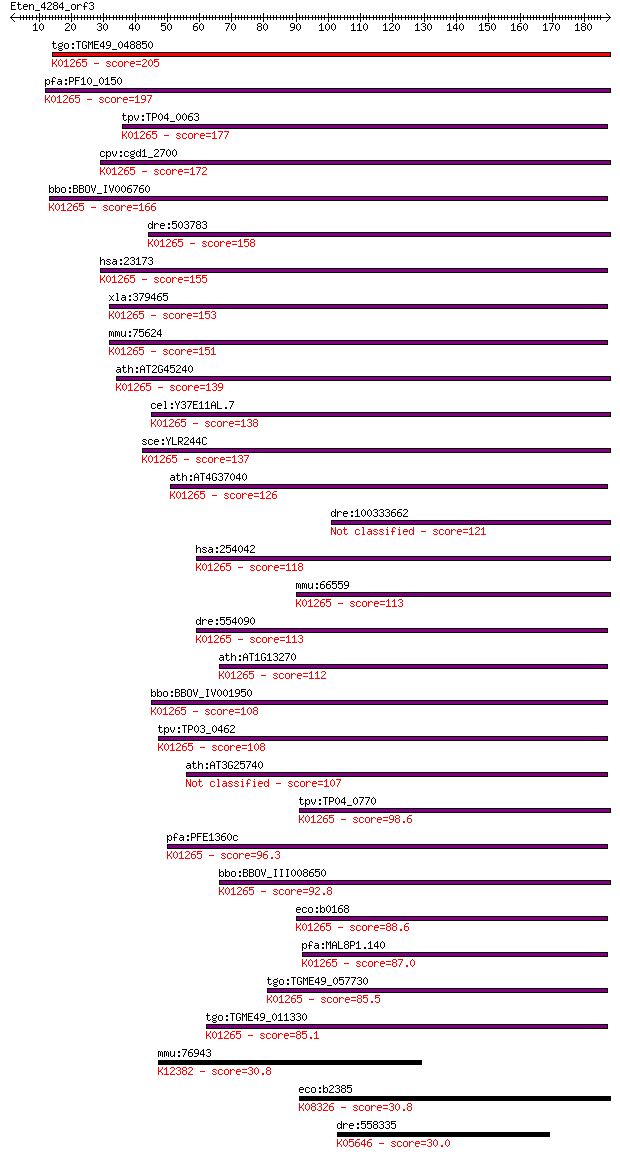
tgo:TGME49_048850 methionine aminopeptidase, putative (EC:3.4.... 205 7e-53
pfa:PF10_0150 methionine aminopeptidase, putative; K01265 meth... 197 1e-50
tpv:TP04_0063 methionine aminopeptidase, type I (EC:3.4.11.18)... 177 1e-44
cpv:cgd1_2700 methionine aminopeptidase with MYND finger at N-... 172 5e-43
bbo:BBOV_IV006760 23.m06237; methionine aminopeptidase (EC:3.4... 166 3e-41
dre:503783 metap1, im:7047238, wu:fc84e12, zgc:110093; methion... 158 9e-39
hsa:23173 METAP1, DKFZp781C0419, KIAA0094, MAP1A, MetAP1A; met... 155 7e-38
xla:379465 metap1-a, MGC64362, metap1; methionyl aminopeptidas... 153 3e-37
mmu:75624 Metap1, 1700029C17Rik, AW047992, KIAA0094, mKIAA0094... 151 1e-36
ath:AT2G45240 MAP1A; MAP1A (METHIONINE AMINOPEPTIDASE 1A); ami... 139 8e-33
cel:Y37E11AL.7 map-1; Methionine AminoPeptidase family member ... 138 1e-32
sce:YLR244C MAP1; Map1p (EC:3.4.11.18); K01265 methionyl amino... 137 3e-32
ath:AT4G37040 MAP1D; MAP1D (METHIONINE AMINOPEPTIDASE 1D); ami... 126 4e-29
dre:100333662 methionyl aminopeptidase 1-like 121 1e-27
hsa:254042 METAP1D, MAP1D, Metap1l; methionyl aminopeptidase t... 118 9e-27
mmu:66559 Metap1d, 2310066F24Rik, 3110033D18Rik, AV117938, Map... 113 3e-25
dre:554090 metap1d, map1d, zgc:110461; methionyl aminopeptidas... 113 5e-25
ath:AT1G13270 MAP1C; MAP1C (METHIONINE AMINOPEPTIDASE 1B); ami... 112 5e-25
bbo:BBOV_IV001950 21.m02884; methionine aminopeptidase I (EC:3... 108 1e-23
tpv:TP03_0462 methionine aminopeptidase, type I (EC:3.4.11.18)... 108 1e-23
ath:AT3G25740 MAP1B; MAP1B (METHIONINE AMINOPEPTIDASE 1C); ami... 107 2e-23
tpv:TP04_0770 methionine aminopeptidase, type I (EC:3.4.11.18)... 98.6 1e-20
pfa:PFE1360c methionine aminopeptidase, putative (EC:3.4.11.18... 96.3 6e-20
bbo:BBOV_III008650 17.m07756; methionine aminopeptidase; K0126... 92.8 7e-19
eco:b0168 map, ECK0166, JW0163, pepM; methionine aminopeptidas... 88.6 1e-17
pfa:MAL8P1.140 methionine aminopeptidase, putative (EC:3.4.11.... 87.0 3e-17
tgo:TGME49_057730 methionine aminopeptidase, putative (EC:3.4.... 85.5 1e-16
tgo:TGME49_011330 methionine aminopeptidase, putative (EC:3.4.... 85.1 1e-16
mmu:76943 Psapl1, 2310020A21Rik; prosaposin-like 1; K12382 sap... 30.8 2.6
eco:b2385 ypdF, ECK2381, JW2382; Xaa-Pro aminopeptidase; K0832... 30.8 2.8
dre:558335 abca12, cb352, sb:cb352; ATP-binding cassette, sub-... 30.0 5.0
> tgo:TGME49_048850 methionine aminopeptidase, putative (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=416
Score = 205 bits (522), Expect = 7e-53, Method: Compositional matrix adjust.
Identities = 100/175 (57%), Positives = 121/175 (69%), Gaps = 3/175 (1%)
Query 14 QTSSSSSSSKGFDPTDVETWKNDPRLREFVGYRFTGALRPWPT-SGPRSVSAGVPLPAYA 72
+TS+ + +K F+P D TW+NDP LR F+ + FTG LRPWP RSV + P YA
Sbjct 77 ETSAETDLAK-FNPQDRNTWRNDPHLRNFLSFSFTGELRPWPILQCMRSVPPHIQQPDYA 135
Query 73 SSAAAAAAAERSFRSCQIKVHNEEEIQRLRAACALGREALDLAHALINPGVTADEIDEKV 132
S + + S R + VH+EEEIQRLR C LGR ALD AH+L+ PGVT +EID KV
Sbjct 136 LSGVPQSELD-SRRKSNVHVHSEEEIQRLRETCLLGRRALDYAHSLVKPGVTTEEIDAKV 194
Query 133 HFFLNTKRAYPSPLNYQGFPKSCCVSVNEVICHGVPDCRPLEKGDIVNVDITVFY 187
H F+ YPSPLNYQ FPKSCC SVNEVICHG+PD RPL+ GDIVN+DITVF+
Sbjct 195 HAFIVDNGGYPSPLNYQQFPKSCCTSVNEVICHGIPDFRPLQDGDIVNIDITVFF 249
> pfa:PF10_0150 methionine aminopeptidase, putative; K01265 methionyl
aminopeptidase [EC:3.4.11.18]
Length=517
Score = 197 bits (502), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 91/176 (51%), Positives = 116/176 (65%), Gaps = 1/176 (0%)
Query 12 LQQTSSSSSSSKGFDPTDVETWKNDPRLREFVGYRFTGALRPWPTSGPRSVSAGVPLPAY 71
L+ S + FDPT+ + W D L+ FV ++FTG +RPWP S V + + P Y
Sbjct 173 LKTIVKKHLSPENFDPTNRKYWVYDDHLKNFVNFKFTGDVRPWPLSKINHVPSHIERPDY 232
Query 72 ASSAAAAAAAERSFRSCQIKVHNEEEIQRLRAACALGREALDLAHALINPGVTADEIDEK 131
A S+ + R I V+NEEEIQR+R AC LGR+ LD AH L++PGVT DEID K
Sbjct 233 AISSIPESELIYK-RKSDIYVNNEEEIQRIREACILGRKTLDYAHTLVSPGVTTDEIDRK 291
Query 132 VHFFLNTKRAYPSPLNYQGFPKSCCVSVNEVICHGVPDCRPLEKGDIVNVDITVFY 187
VH F+ AYPS LNY FPKSCC SVNE++CHG+PD RPL+ GDI+N+DI+VFY
Sbjct 292 VHEFIIKNNAYPSTLNYYKFPKSCCTSVNEIVCHGIPDYRPLKSGDIINIDISVFY 347
> tpv:TP04_0063 methionine aminopeptidase, type I (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=378
Score = 177 bits (450), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 81/151 (53%), Positives = 106/151 (70%), Gaps = 1/151 (0%)
Query 36 DPRLREFVGYRFTGALRPWPTSGPRSVSAGVPLPAYASSAAAAAAAERSFRSCQIKVHNE 95
D L++F ++FTG LRPWP + + V +P P YA + + S IKVH+
Sbjct 65 DFYLKKFRNFKFTGDLRPWPVTDMKKVPKHIPRPDYAEDGIPHSEINEKY-SNAIKVHDP 123
Query 96 EEIQRLRAACALGREALDLAHALINPGVTADEIDEKVHFFLNTKRAYPSPLNYQGFPKSC 155
+ I+++R AC LGR+ALDLA++LI PG+T DEID KVH F+ + YPSPLNY FPKS
Sbjct 124 QTIKKIRRACLLGRKALDLANSLIKPGITTDEIDTKVHEFIVSHNGYPSPLNYYNFPKSI 183
Query 156 CVSVNEVICHGVPDCRPLEKGDIVNVDITVF 186
C SVNEV+CHG+PD RPLE+GDIVNVDI+V+
Sbjct 184 CTSVNEVVCHGIPDLRPLEEGDIVNVDISVY 214
> cpv:cgd1_2700 methionine aminopeptidase with MYND finger at
N-terminus ; K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=407
Score = 172 bits (437), Expect = 5e-43, Method: Compositional matrix adjust.
Identities = 85/161 (52%), Positives = 108/161 (67%), Gaps = 2/161 (1%)
Query 29 DVETWKNDPRLREFVGYR-FTGALRPWPTSGPRSVSAGVPLPAYASSAAAAAAAE-RSFR 86
D TW N + +F+G+ FTG LRP+P S R V + + P YA +E + +
Sbjct 84 DPRTWVNCSHISKFIGFNGFTGPLRPYPISIKRKVPSHILRPDYADDKEGRPFSELKRKK 143
Query 87 SCQIKVHNEEEIQRLRAACALGREALDLAHALINPGVTADEIDEKVHFFLNTKRAYPSPL 146
S I EEI+ LR C +GREALD+A ++I PGVT D IDE VH F+ +K +YPSPL
Sbjct 144 SSAIVTATAEEIELLRECCKIGREALDIAASMIKPGVTTDAIDEAVHNFIISKNSYPSPL 203
Query 147 NYQGFPKSCCVSVNEVICHGVPDCRPLEKGDIVNVDITVFY 187
NY FPKSCC SVNE+ICHG+PD RPLE+GDIVNVDI+V+Y
Sbjct 204 NYWEFPKSCCTSVNEIICHGIPDFRPLEEGDIVNVDISVYY 244
> bbo:BBOV_IV006760 23.m06237; methionine aminopeptidase (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=376
Score = 166 bits (421), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 84/174 (48%), Positives = 110/174 (63%), Gaps = 8/174 (4%)
Query 13 QQTSSSSSSSKGFDPTDVETWKNDPRLREFVGYRFTGALRPWPTSGPRSVSAGVPLPAYA 72
++ +S S SS D NDP LR F ++FTG LRPWP + + V + + P YA
Sbjct 50 KEDTSGSVSSGDLD-------LNDPHLRRFKNFKFTGELRPWPVTPQKRVPSHISCPDYA 102
Query 73 SSAAAAAAAERSFRSCQIKVHNEEEIQRLRAACALGREALDLAHALINPGVTADEIDEKV 132
+ + +I V+ E+I+ +R A LGR+ALD A +LI PGVT DEID KV
Sbjct 103 LDGEPKSEVNLK-NAGRIVVNTPEQIKLIRKASILGRKALDFAASLIAPGVTTDEIDTKV 161
Query 133 HFFLNTKRAYPSPLNYQGFPKSCCVSVNEVICHGVPDCRPLEKGDIVNVDITVF 186
H F+ AYPSPLNY GFPKS C SVNEV+CHG+PD RPL+ GDI+N+DI+V+
Sbjct 162 HDFIIQHNAYPSPLNYYGFPKSLCTSVNEVVCHGIPDKRPLKDGDIINIDISVY 215
> dre:503783 metap1, im:7047238, wu:fc84e12, zgc:110093; methionyl
aminopeptidase 1 (EC:3.4.11.18); K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=386
Score = 158 bits (400), Expect = 9e-39, Method: Compositional matrix adjust.
Identities = 73/146 (50%), Positives = 99/146 (67%), Gaps = 2/146 (1%)
Query 44 GYRFTGALRPW-PTSGPRSVSAGVPLPAYASSAAAAAAAERSFR-SCQIKVHNEEEIQRL 101
GYR+TG LRP+ P + R V + + P YA + +E++ + + QIK+ N EEI+ +
Sbjct 79 GYRYTGKLRPYYPLTPMRLVPSNIQRPDYADHPLGMSESEQTMKGTSQIKILNAEEIEGM 138
Query 102 RAACALGREALDLAHALINPGVTADEIDEKVHFFLNTKRAYPSPLNYQGFPKSCCVSVNE 161
R C L RE LD+A ++ PGVT +EID VH + YPSPLNY FPKSCC SVNE
Sbjct 139 RVVCKLAREVLDIAAMMVKPGVTTEEIDHAVHLACTARNCYPSPLNYYNFPKSCCTSVNE 198
Query 162 VICHGVPDCRPLEKGDIVNVDITVFY 187
VICHG+PD R L++GDI+N+DITV++
Sbjct 199 VICHGIPDRRHLQEGDILNIDITVYH 224
> hsa:23173 METAP1, DKFZp781C0419, KIAA0094, MAP1A, MetAP1A; methionyl
aminopeptidase 1 (EC:3.4.11.18); K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=386
Score = 155 bits (392), Expect = 7e-38, Method: Compositional matrix adjust.
Identities = 77/162 (47%), Positives = 105/162 (64%), Gaps = 4/162 (2%)
Query 29 DVETW--KNDPRLREFVGYRFTGALRP-WPTSGPRSVSAGVPLPAYASSAAAAAAAERSF 85
+V +W + D + GYR+TG LRP +P R V + + P YA + +E++
Sbjct 63 EVSSWTVEGDINTDPWAGYRYTGKLRPHYPLMPTRPVPSYIQRPDYADHPLGMSESEQAL 122
Query 86 R-SCQIKVHNEEEIQRLRAACALGREALDLAHALINPGVTADEIDEKVHFFLNTKRAYPS 144
+ + QIK+ + E+I+ +R C L RE LD+A +I PGVT +EID VH + YPS
Sbjct 123 KGTSQIKLLSSEDIEGMRLVCRLAREVLDVAAGMIKPGVTTEEIDHAVHLACIARNCYPS 182
Query 145 PLNYQGFPKSCCVSVNEVICHGVPDCRPLEKGDIVNVDITVF 186
PLNY FPKSCC SVNEVICHG+PD RPL++GDIVNVDIT++
Sbjct 183 PLNYYNFPKSCCTSVNEVICHGIPDRRPLQEGDIVNVDITLY 224
> xla:379465 metap1-a, MGC64362, metap1; methionyl aminopeptidase
1 (EC:3.4.11.18); K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=385
Score = 153 bits (386), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 74/157 (47%), Positives = 102/157 (64%), Gaps = 2/157 (1%)
Query 32 TWKNDPRLREFVGYRFTGALRP-WPTSGPRSVSAGVPLPAYASSAAAAAAAERSFR-SCQ 89
T +D + GYR+TG LRP +P + R V+ + P YA + +E++ + + Q
Sbjct 68 TMDDDVNTDPWPGYRYTGKLRPHYPLTPMRPVTNHIQRPDYADHPLGMSESEQTLKGTSQ 127
Query 90 IKVHNEEEIQRLRAACALGREALDLAHALINPGVTADEIDEKVHFFLNTKRAYPSPLNYQ 149
IKV + E+I+ +R C L RE L +A ++ PG+T +EID VH ++ YPSPLNY
Sbjct 128 IKVLSPEDIEGMRVVCRLAREVLGVAAMMVKPGITTEEIDHAVHLACISRSCYPSPLNYY 187
Query 150 GFPKSCCVSVNEVICHGVPDCRPLEKGDIVNVDITVF 186
FPKSCC SVNE+ICHG+PD RPL+ GDIVNVDITV+
Sbjct 188 NFPKSCCTSVNEIICHGIPDRRPLQDGDIVNVDITVY 224
> mmu:75624 Metap1, 1700029C17Rik, AW047992, KIAA0094, mKIAA0094;
methionyl aminopeptidase 1 (EC:3.4.11.18); K01265 methionyl
aminopeptidase [EC:3.4.11.18]
Length=386
Score = 151 bits (382), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 75/157 (47%), Positives = 101/157 (64%), Gaps = 2/157 (1%)
Query 32 TWKNDPRLREFVGYRFTGALRP-WPTSGPRSVSAGVPLPAYASSAAAAAAAERSFR-SCQ 89
T + D + GYR+TG LRP +P R V + + P YA + +E++ + + Q
Sbjct 68 TVEGDVNTDPWAGYRYTGKLRPHYPLMPTRPVPSYIQRPDYADHPLGMSESEQALKGTSQ 127
Query 90 IKVHNEEEIQRLRAACALGREALDLAHALINPGVTADEIDEKVHFFLNTKRAYPSPLNYQ 149
IK+ + E+I+ +R C L RE LD+A +I GVT +EID VH + YPSPLNY
Sbjct 128 IKLLSSEDIEGMRLVCRLAREVLDIAAGMIKAGVTTEEIDHAVHLACIARNCYPSPLNYY 187
Query 150 GFPKSCCVSVNEVICHGVPDCRPLEKGDIVNVDITVF 186
FPKSCC SVNEVICHG+PD RPL++GDIVNVDIT++
Sbjct 188 NFPKSCCTSVNEVICHGIPDRRPLQEGDIVNVDITLY 224
> ath:AT2G45240 MAP1A; MAP1A (METHIONINE AMINOPEPTIDASE 1A); aminopeptidase/
metalloexopeptidase; K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=398
Score = 139 bits (349), Expect = 8e-33, Method: Compositional matrix adjust.
Identities = 71/154 (46%), Positives = 89/154 (57%), Gaps = 1/154 (0%)
Query 34 KNDPRLREFVGYRFTGALRPWPTSGPRSVSAGVPLPAYASSAAAAAAAERSFRSCQIKVH 93
K R + + +TG L+ +P S R V A + P +A + +++
Sbjct 84 KGQARTPKLPHFDWTGPLKQYPISTKRVVPAEIEKPDWAIDGTPKVEPNSDLQHV-VEIK 142
Query 94 NEEEIQRLRAACALGREALDLAHALINPGVTADEIDEKVHFFLNTKRAYPSPLNYQGFPK 153
E+IQR+R C + RE LD A +I+PGVT DEID VH YPSPLNY FPK
Sbjct 143 TPEQIQRMRETCKIAREVLDAAARVIHPGVTTDEIDRVVHEATIAAGGYPSPLNYYFFPK 202
Query 154 SCCVSVNEVICHGVPDCRPLEKGDIVNVDITVFY 187
SCC SVNEVICHG+PD R LE GDIVNVD+TV Y
Sbjct 203 SCCTSVNEVICHGIPDARKLEDGDIVNVDVTVCY 236
> cel:Y37E11AL.7 map-1; Methionine AminoPeptidase family member
(map-1); K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=371
Score = 138 bits (347), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 72/144 (50%), Positives = 89/144 (61%), Gaps = 1/144 (0%)
Query 45 YRFTGALRPWPTSGPRSVSAGVPLPAYASSAAAAAAAERSFRSCQ-IKVHNEEEIQRLRA 103
Y FTG+LRP + R V +P P YA + ER +S + IKV +EE + L+
Sbjct 63 YSFTGSLRPGRVTDRRPVPDHIPRPDYALHPQGVSLEERQSKSERVIKVLTDEEKEGLKV 122
Query 104 ACALGREALDLAHALINPGVTADEIDEKVHFFLNTKRAYPSPLNYQGFPKSCCVSVNEVI 163
AC LGRE L+ A PGVT +EID VH + YPSPL Y FPKSCC SVNEVI
Sbjct 123 ACKLGRECLNEAAKACGPGVTTEEIDRVVHEAAIERDCYPSPLGYYKFPKSCCTSVNEVI 182
Query 164 CHGVPDCRPLEKGDIVNVDITVFY 187
CHG+PD R LE GD+ NVD+TV++
Sbjct 183 CHGIPDMRKLENGDLCNVDVTVYH 206
> sce:YLR244C MAP1; Map1p (EC:3.4.11.18); K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=387
Score = 137 bits (344), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 65/147 (44%), Positives = 100/147 (68%), Gaps = 2/147 (1%)
Query 42 FVGYRFTGALR-PWPTSGPRSVSAGVPLPAYASSAAAAAAAERSFRSCQIKVHNEEEIQR 100
F ++++G ++ +P + R V +P P +A++ + +R+ R I ++ +++I++
Sbjct 79 FPKFKYSGKVKASYPLTPRRYVPEDIPKPDWAANGLPVSE-QRNDRLNNIPIYKKDQIKK 137
Query 101 LRAACALGREALDLAHALINPGVTADEIDEKVHFFLNTKRAYPSPLNYQGFPKSCCVSVN 160
+R AC LGRE LD+A A + PG+T DE+DE VH + AYPSPLNY FPKS C SVN
Sbjct 138 IRKACMLGREVLDIAAAHVRPGITTDELDEIVHNETIKRGAYPSPLNYYNFPKSLCTSVN 197
Query 161 EVICHGVPDCRPLEKGDIVNVDITVFY 187
EVICHGVPD L++GDIVN+D++++Y
Sbjct 198 EVICHGVPDKTVLKEGDIVNLDVSLYY 224
> ath:AT4G37040 MAP1D; MAP1D (METHIONINE AMINOPEPTIDASE 1D); aminopeptidase/
metalloexopeptidase; K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=350
Score = 126 bits (317), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 65/136 (47%), Positives = 81/136 (59%), Gaps = 6/136 (4%)
Query 51 LRPWPTSGPRSVSAGVPLPAYASSAAAAAAAERSFRSCQIKVHNEEEIQRLRAACALGRE 110
LRP S R V + P Y S A S ++VH+++ I+ +RA+ L
Sbjct 72 LRPGNVSPRRPVPGHITKPPYVDSLQAPGI------SSGLEVHDKKGIECMRASGILAAR 125
Query 111 ALDLAHALINPGVTADEIDEKVHFFLNTKRAYPSPLNYQGFPKSCCVSVNEVICHGVPDC 170
D A L+ PGVT DEIDE VH + AYPSPL Y GFPKS C SVNE ICHG+PD
Sbjct 126 VRDYAGTLVKPGVTTDEIDEAVHNMIIENGAYPSPLGYGGFPKSVCTSVNECICHGIPDS 185
Query 171 RPLEKGDIVNVDITVF 186
RPLE GDI+N+D+TV+
Sbjct 186 RPLEDGDIINIDVTVY 201
> dre:100333662 methionyl aminopeptidase 1-like
Length=249
Score = 121 bits (303), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 51/87 (58%), Positives = 64/87 (73%), Gaps = 0/87 (0%)
Query 101 LRAACALGREALDLAHALINPGVTADEIDEKVHFFLNTKRAYPSPLNYQGFPKSCCVSVN 160
+R C L RE LD+A ++ PGVT +EID VH + YPSPLNY FPKSCC SVN
Sbjct 1 MRVVCKLAREVLDIAAMMVKPGVTTEEIDHAVHLACTARNCYPSPLNYYNFPKSCCTSVN 60
Query 161 EVICHGVPDCRPLEKGDIVNVDITVFY 187
EVICHG+PD R L++GDI+N+DITV++
Sbjct 61 EVICHGIPDRRHLQEGDILNIDITVYH 87
> hsa:254042 METAP1D, MAP1D, Metap1l; methionyl aminopeptidase
type 1D (mitochondrial) (EC:3.4.11.18); K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=335
Score = 118 bits (296), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 58/129 (44%), Positives = 77/129 (59%), Gaps = 0/129 (0%)
Query 59 PRSVSAGVPLPAYASSAAAAAAAERSFRSCQIKVHNEEEIQRLRAACALGREALDLAHAL 118
P +VS+ P+P + I+V NE++IQ L AC L R L LA
Sbjct 55 PAAVSSAHPVPKHIKKPDYVTTGIVPDWGDSIEVKNEDQIQGLHQACQLARHVLLLAGKS 114
Query 119 INPGVTADEIDEKVHFFLNTKRAYPSPLNYQGFPKSCCVSVNEVICHGVPDCRPLEKGDI 178
+ +T +EID VH + + AYPSPL Y GFPKS C SVN V+CHG+PD RPL+ GDI
Sbjct 115 LKVDMTTEEIDALVHREIISHNAYPSPLGYGGFPKSVCTSVNNVLCHGIPDSRPLQDGDI 174
Query 179 VNVDITVFY 187
+N+D+TV+Y
Sbjct 175 INIDVTVYY 183
> mmu:66559 Metap1d, 2310066F24Rik, 3110033D18Rik, AV117938, Map1d,
Metapl1; methionyl aminopeptidase type 1D (mitochondrial)
(EC:3.4.11.18); K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=335
Score = 113 bits (283), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 52/98 (53%), Positives = 68/98 (69%), Gaps = 0/98 (0%)
Query 90 IKVHNEEEIQRLRAACALGREALDLAHALINPGVTADEIDEKVHFFLNTKRAYPSPLNYQ 149
I+V +E++IQ LR AC L R L LA + +T +EID VH+ + AYPSPL Y
Sbjct 86 IEVKDEDQIQGLREACRLARHVLLLAGKSLKVDMTTEEIDALVHWEIIRHDAYPSPLGYG 145
Query 150 GFPKSCCVSVNEVICHGVPDCRPLEKGDIVNVDITVFY 187
FPKS C SVN V+CHG+PD RPL+ GDI+N+D+TV+Y
Sbjct 146 RFPKSVCTSVNNVLCHGIPDSRPLQDGDIINIDVTVYY 183
> dre:554090 metap1d, map1d, zgc:110461; methionyl aminopeptidase
type 1D (mitochondrial) (EC:3.4.11.18); K01265 methionyl
aminopeptidase [EC:3.4.11.18]
Length=338
Score = 113 bits (282), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 56/128 (43%), Positives = 76/128 (59%), Gaps = 0/128 (0%)
Query 59 PRSVSAGVPLPAYASSAAAAAAAERSFRSCQIKVHNEEEIQRLRAACALGREALDLAHAL 118
P V P+P + ++++ I++ +EE+IQ LR AC L R L L
Sbjct 58 PAIVRPAYPVPKHIQRPDYVSSSKVPEWPDYIEIKDEEQIQGLRRACQLARHILLLTGNS 117
Query 119 INPGVTADEIDEKVHFFLNTKRAYPSPLNYQGFPKSCCVSVNEVICHGVPDCRPLEKGDI 178
+ G+T DEID VH YPSPL+Y GFPKS C SVN V+CHG+PD RPL+ GDI
Sbjct 118 LKVGMTTDEIDFIVHQEAIRHNGYPSPLHYGGFPKSVCTSVNNVVCHGIPDSRPLQDGDI 177
Query 179 VNVDITVF 186
+N+D+TV+
Sbjct 178 INIDVTVY 185
> ath:AT1G13270 MAP1C; MAP1C (METHIONINE AMINOPEPTIDASE 1B); aminopeptidase/
metalloexopeptidase; K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=369
Score = 112 bits (281), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 54/121 (44%), Positives = 72/121 (59%), Gaps = 6/121 (4%)
Query 66 VPLPAYASSAAAAAAAERSFRSCQIKVHNEEEIQRLRAACALGREALDLAHALINPGVTA 125
+P P Y S S + ++ E I ++RAAC L L+ A L+ P VT
Sbjct 106 IPRPPYVESGVLPDI------SSEFQIPGPEGIAKMRAACELAARVLNYAGTLVKPSVTT 159
Query 126 DEIDEKVHFFLNTKRAYPSPLNYQGFPKSCCVSVNEVICHGVPDCRPLEKGDIVNVDITV 185
+EID+ VH + AYPSPL Y GFPKS C SVNE +CHG+PD R L+ GDI+N+D+TV
Sbjct 160 NEIDKAVHDMIIEAGAYPSPLGYGGFPKSVCTSVNECMCHGIPDSRQLQSGDIINIDVTV 219
Query 186 F 186
+
Sbjct 220 Y 220
> bbo:BBOV_IV001950 21.m02884; methionine aminopeptidase I (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=611
Score = 108 bits (269), Expect = 1e-23, Method: Composition-based stats.
Identities = 56/148 (37%), Positives = 76/148 (51%), Gaps = 6/148 (4%)
Query 45 YRFTGALRPWPTSGPRSVSAGVPLPAYASSAAAAAAAE------RSFRSCQIKVHNEEEI 98
Y FTG ++ +G + P Y +S R + + + EI
Sbjct 132 YEFTGPIKKGIVTGRLLPGKNIKRPNYYASGLPKYVDYPRDVDYRGEHDPRGTIKTDSEI 191
Query 99 QRLRAACALGREALDLAHALINPGVTADEIDEKVHFFLNTKRAYPSPLNYQGFPKSCCVS 158
+R AC + RE LD LI G+ D ID +H K+ YPSPLNY GFPKS C S
Sbjct 192 AGIRYACRIAREVLDSVSPLIVEGMFTDYIDRHIHRMCRLKKVYPSPLNYHGFPKSVCTS 251
Query 159 VNEVICHGVPDCRPLEKGDIVNVDITVF 186
+NE++CHG+PD L GDI+NVD+TV+
Sbjct 252 INEIVCHGIPDSTVLAAGDIINVDVTVY 279
> tpv:TP03_0462 methionine aminopeptidase, type I (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=461
Score = 108 bits (269), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 52/144 (36%), Positives = 78/144 (54%), Gaps = 4/144 (2%)
Query 47 FTGALRPWPTSGPRSVSAGVPLPAYASSAAAAAA----AERSFRSCQIKVHNEEEIQRLR 102
FTG +R SG + S + P Y + R + + + N+E+I ++
Sbjct 2 FTGHIRKGVLSGKQYPSKNIKRPNYFKTGKPTYVDYPYCNRDAKDIRGTIKNDEDIAGIK 61
Query 103 AACALGREALDLAHALINPGVTADEIDEKVHFFLNTKRAYPSPLNYQGFPKSCCVSVNEV 162
AC + RE LD +++ G+ D+ID VH K+ +P+ LNY GFPKS C S+NEV
Sbjct 62 NACRIAREILDYVSSIVVEGIFTDDIDRFVHKLCAIKKVFPATLNYHGFPKSVCTSINEV 121
Query 163 ICHGVPDCRPLEKGDIVNVDITVF 186
CHG+PD L GD++ VD+TV+
Sbjct 122 ACHGIPDNNLLCAGDLLKVDLTVY 145
> ath:AT3G25740 MAP1B; MAP1B (METHIONINE AMINOPEPTIDASE 1C); aminopeptidase/
metalloexopeptidase
Length=344
Score = 107 bits (267), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 56/132 (42%), Positives = 76/132 (57%), Gaps = 7/132 (5%)
Query 56 TSGPR-SVSAGVPLPAYASSAAAAAAAERSFRSCQIKVHNEEEIQRLRAACALGREALDL 114
T PR SV + P Y S+ S ++++ + I +++ AC L LD
Sbjct 68 TVSPRLSVPDHILKPLYVESSKVPEI------SSELQIPDSIGIVKMKKACELAARVLDY 121
Query 115 AHALINPGVTADEIDEKVHFFLNTKRAYPSPLNYQGFPKSCCVSVNEVICHGVPDCRPLE 174
A L+ P VT DEID+ VH + AYPSPL Y GFPKS C SVNE + HG+PD RPL+
Sbjct 122 AGTLVRPFVTTDEIDKAVHQMVIEFGAYPSPLGYGGFPKSVCTSVNECMFHGIPDSRPLQ 181
Query 175 KGDIVNVDITVF 186
GDI+N+D+ V+
Sbjct 182 NGDIINIDVAVY 193
> tpv:TP04_0770 methionine aminopeptidase, type I (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=293
Score = 98.6 bits (244), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 45/97 (46%), Positives = 64/97 (65%), Gaps = 0/97 (0%)
Query 91 KVHNEEEIQRLRAACALGREALDLAHALINPGVTADEIDEKVHFFLNTKRAYPSPLNYQG 150
+V NEE+I ++RAA + + L L GVTAD +D K F+ + AYPS +N+ G
Sbjct 87 EVKNEEQISKMRAAAKIAAKCLKLCLDSTEKGVTADFVDRKAQDFIVSSGAYPSGVNFHG 146
Query 151 FPKSCCVSVNEVICHGVPDCRPLEKGDIVNVDITVFY 187
+P++ CVSVNEV CHG+P+ RP + D+V+ D TVFY
Sbjct 147 YPRAICVSVNEVACHGIPNMRPFHESDVVSYDCTVFY 183
> pfa:PFE1360c methionine aminopeptidase, putative (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=327
Score = 96.3 bits (238), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 46/137 (33%), Positives = 71/137 (51%), Gaps = 0/137 (0%)
Query 50 ALRPWPTSGPRSVSAGVPLPAYASSAAAAAAAERSFRSCQIKVHNEEEIQRLRAACALGR 109
+P G V+A +P + + A + ++ +++ I++++ AC L
Sbjct 42 TFKPTLQDGKYKVTASQKVPEHIKCPSYAKTGIVQNSNINYEIKDDKYIEKMKKACKLAS 101
Query 110 EALDLAHALINPGVTADEIDEKVHFFLNTKRAYPSPLNYQGFPKSCCVSVNEVICHGVPD 169
E L L G+T D ID + F AYP+ LN+ FPKS C S NEV+CHG+P+
Sbjct 102 ECLKLCLENSKEGITTDIIDNMAYDFYIKNNAYPAGLNFHSFPKSICASPNEVVCHGIPN 161
Query 170 CRPLEKGDIVNVDITVF 186
R L++GDI+ D TVF
Sbjct 162 LRKLQRGDIITYDCTVF 178
> bbo:BBOV_III008650 17.m07756; methionine aminopeptidase; K01265
methionyl aminopeptidase [EC:3.4.11.18]
Length=330
Score = 92.8 bits (229), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 44/123 (35%), Positives = 75/123 (60%), Gaps = 1/123 (0%)
Query 66 VPLPAYASSAAAAAAAERSFRSCQI-KVHNEEEIQRLRAACALGREALDLAHALINPGVT 124
+ LP YAS + ++C +V ++ +I +RAA + L + GVT
Sbjct 57 IALPPYASIRDPSELRAYYNKACTFAEVKSQSQIALMRAAAKIAAGCLKHCINITKEGVT 116
Query 125 ADEIDEKVHFFLNTKRAYPSPLNYQGFPKSCCVSVNEVICHGVPDCRPLEKGDIVNVDIT 184
A++ID + H ++ ++ +YP+ + + GFPK+ C+S+NEV CHG+P+ RP ++GDIV+ D T
Sbjct 117 AEDIDREGHEYIVSQGSYPAGVGFHGFPKAICISINEVACHGIPNTRPFQRGDIVSYDCT 176
Query 185 VFY 187
VF+
Sbjct 177 VFH 179
> eco:b0168 map, ECK0166, JW0163, pepM; methionine aminopeptidase
(EC:3.4.11.18); K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=264
Score = 88.6 bits (218), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 40/99 (40%), Positives = 64/99 (64%), Gaps = 2/99 (2%)
Query 90 IKVHNEEEIQRLRAACALGREALDLAHALINPGVTADEIDEKVH-FFLNTKRAYPSPLNY 148
I + E+I+++R A L E L++ + PGV+ E+D + + +N + A + L Y
Sbjct 3 ISIKTPEDIEKMRVAGRLAAEVLEMIEPYVKPGVSTGELDRICNDYIVNEQHAVSACLGY 62
Query 149 QGFPKSCCVSVNEVICHGVP-DCRPLEKGDIVNVDITVF 186
G+PKS C+S+NEV+CHG+P D + L+ GDIVN+D+TV
Sbjct 63 HGYPKSVCISINEVVCHGIPDDAKLLKDGDIVNIDVTVI 101
> pfa:MAL8P1.140 methionine aminopeptidase, putative (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=740
Score = 87.0 bits (214), Expect = 3e-17, Method: Composition-based stats.
Identities = 37/95 (38%), Positives = 59/95 (62%), Gaps = 0/95 (0%)
Query 92 VHNEEEIQRLRAACALGREALDLAHALINPGVTADEIDEKVHFFLNTKRAYPSPLNYQGF 151
+ ++++IQ ++ C RE +D +I G+T ++ID + YPSPLNY F
Sbjct 264 IKSDKDIQIIKENCKFARELMDDVSYIICEGITTNDIDIYILNKCINNGFYPSPLNYHNF 323
Query 152 PKSCCVSVNEVICHGVPDCRPLEKGDIVNVDITVF 186
PKS C+S+NE++CHG+PD L D+V +DI++F
Sbjct 324 PKSSCISINEILCHGIPDNNLLYLNDVVKIDISLF 358
> tgo:TGME49_057730 methionine aminopeptidase, putative (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=387
Score = 85.5 bits (210), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 42/108 (38%), Positives = 67/108 (62%), Gaps = 2/108 (1%)
Query 81 AERSFRSCQI--KVHNEEEIQRLRAACALGREALDLAHALINPGVTADEIDEKVHFFLNT 138
A+R F + +V + ++++RAA + AL L GVT +++D+ VH ++ +
Sbjct 128 ADRGFDTSDTPGEVQTPDALKKIRAAATVAANALKLGLDAAREGVTTEDLDKIVHEYIVS 187
Query 139 KRAYPSPLNYQGFPKSCCVSVNEVICHGVPDCRPLEKGDIVNVDITVF 186
AYP+ +N+ FPK+ C SVNE +CHG+PD RPL+ GDIV +D T +
Sbjct 188 VGAYPAAVNFHNFPKAVCASVNEAVCHGIPDLRPLQDGDIVTLDCTAY 235
> tgo:TGME49_011330 methionine aminopeptidase, putative (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=329
Score = 85.1 bits (209), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 54/160 (33%), Positives = 79/160 (49%), Gaps = 36/160 (22%)
Query 62 VSAGVPLPAYA--------SSAAAAAAAERSFRSCQ---IK----------------VHN 94
V AG+ P YA + A +AE R C+ +K +
Sbjct 4 VGAGIVKPHYAEEDDGRAKAKREAMLSAELQLRRCEGDEMKARLARFHPVAEREMEWIKP 63
Query 95 EEEIQRLRAACALGREALDLAHALINPGV--------TADEIDEKVHFFLNTKRAYPSPL 146
+ E++ +R AC + RE L +A + GV T ++ID VH + AYPSPL
Sbjct 64 QAEVEGVRRACEVTREVLQVAVDFVK-GVCAQSSAPLTTEDIDRVVHEETMKRGAYPSPL 122
Query 147 NYQGFPKSCCVSVNEVICHGVPDCRPLEKGDIVNVDITVF 186
Y FPKS C S NE++CHG+PD RPL++G I ++D++ F
Sbjct 123 RYCNFPKSVCTSTNEIVCHGIPDDRPLQRGSICSIDVSCF 162
> mmu:76943 Psapl1, 2310020A21Rik; prosaposin-like 1; K12382 saposin
Length=525
Score = 30.8 bits (68), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 37/82 (45%), Gaps = 5/82 (6%)
Query 47 FTGALRPWPTSGPRSVSAGVPLPAYASSAAAAAAAERSFRSCQIKVHNEEEIQRLRAACA 106
GA R P S PR + G + AAA R+ R CQ V N+ ++ L C+
Sbjct 14 LLGAARASPISVPRECAKGSEVWCQDLQAAAKC---RAVRHCQSAVWNKPTVKSL--PCS 68
Query 107 LGREALDLAHALINPGVTADEI 128
+ ++ A +NPG T +I
Sbjct 69 VCQDVAAAAGNGVNPGATESDI 90
> eco:b2385 ypdF, ECK2381, JW2382; Xaa-Pro aminopeptidase; K08326
aminopeptidase [EC:3.4.11.-]
Length=361
Score = 30.8 bits (68), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 18/97 (18%), Positives = 39/97 (40%), Gaps = 5/97 (5%)
Query 91 KVHNEEEIQRLRAACALGREALDLAHALINPGVTADEIDEKVHFFLNTKRAYPSPLNYQG 150
++ EE++++R AC + + I G++ EI ++ +F+ + A +
Sbjct 126 QIKTPEEVEKIRLACGIADRGAEHIRRFIQAGMSEREIAAELEWFMRQQGAEKAS----- 180
Query 151 FPKSCCVSVNEVICHGVPDCRPLEKGDIVNVDITVFY 187
F + HG + + G+ V +D Y
Sbjct 181 FDTIVASGWRGALPHGKASDKIVAAGEFVTLDFGALY 217
> dre:558335 abca12, cb352, sb:cb352; ATP-binding cassette, sub-family
A (ABC1), member 12; K05646 ATP-binding cassette, subfamily
A (ABC1), member 12
Length=3644
Score = 30.0 bits (66), Expect = 5.0, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 32/73 (43%), Gaps = 9/73 (12%)
Query 103 AACALGREALDLAH---ALINPGVTADEIDEKVHFFLN----TKRAYPSPLNYQGFPKSC 155
AA LG DL H +++P + +DE+ FF N T R + ++Y G C
Sbjct 2807 AAMGLGSIKSDLQHFPEIVLSPALY--HVDEQYAFFSNQNPNTSRLVDTMMSYPGIDHVC 2864
Query 156 CVSVNEVICHGVP 168
+ +C G P
Sbjct 2865 MTDPTDPVCKGRP 2877
Lambda K H
0.317 0.132 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5235419772
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40