bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4296_orf2
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
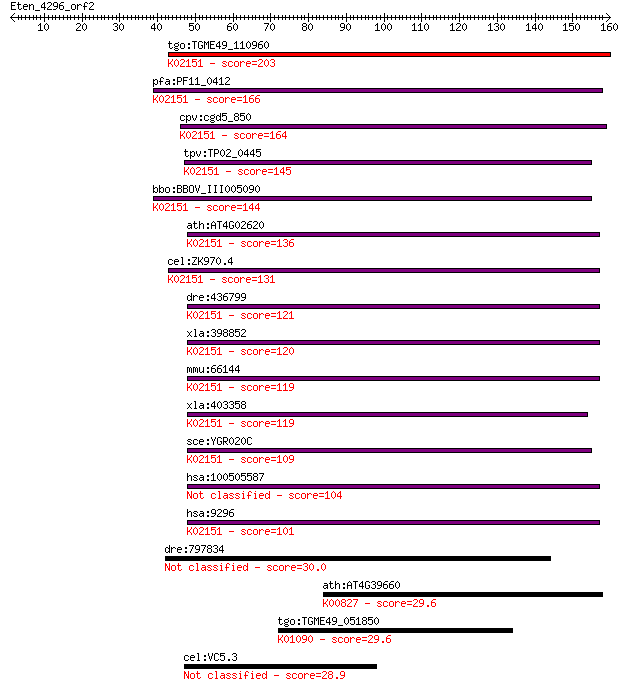
tgo:TGME49_110960 vacuolar ATP synthase subunit F, putative (E... 203 2e-52
pfa:PF11_0412 vacuolar ATP synthase subunit F, putative; K0215... 166 3e-41
cpv:cgd5_850 vacuolar ATP synthase subunit F ; K02151 V-type H... 164 1e-40
tpv:TP02_0445 vacuolar ATP synthase subunit F; K02151 V-type H... 145 8e-35
bbo:BBOV_III005090 17.m07454; vacuolar ATP synthase subunit F;... 144 2e-34
ath:AT4G02620 vacuolar ATPase subunit F family protein; K02151... 136 4e-32
cel:ZK970.4 vha-9; Vacuolar H ATPase family member (vha-9); K0... 131 1e-30
dre:436799 atp6v1f, zgc:92923; ATPase, H+ transporting, V1 sub... 121 1e-27
xla:398852 atp6v1f-b, MGC115045, MGC68786, Vma7, atp6s14, atp6... 120 2e-27
mmu:66144 Atp6v1f, 1110004G16Rik; ATPase, H+ transporting, lys... 119 6e-27
xla:403358 hypothetical protein MGC68592; K02151 V-type H+-tra... 119 6e-27
sce:YGR020C VMA7; Subunit F of the eight-subunit V1 peripheral... 109 3e-24
hsa:100505587 v-type proton ATPase subunit F-like 104 1e-22
hsa:9296 ATP6V1F, ATP6S14, MGC117321, MGC126037, MGC126038, VA... 101 8e-22
dre:797834 cb606, sb:cb606; si:dkeyp-110c7.1 30.0 3.4
ath:AT4G39660 AGT2; AGT2 (ALANINE:GLYOXYLATE AMINOTRANSFERASE ... 29.6 4.0
tgo:TGME49_051850 serine/threonine protein phosphatase 5, puta... 29.6 4.0
cel:VC5.3 npa-1; Nematode Polyprotein Allergen related family ... 28.9 7.5
> tgo:TGME49_110960 vacuolar ATP synthase subunit F, putative
(EC:3.6.3.14); K02151 V-type H+-transporting ATPase subunit
F [EC:3.6.3.14]
Length=127
Score = 203 bits (516), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 90/117 (76%), Positives = 107/117 (91%), Gaps = 0/117 (0%)
Query 43 AAELKICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHRSDVG 102
+LK+ VIGDEDTVAGFLMAGIGMRDG+G+TNF +VD+KTKRQD+E AF T R D+G
Sbjct 10 GTDLKVAVIGDEDTVAGFLMAGIGMRDGLGRTNFFIVDSKTKRQDVEDAFRTMTERPDIG 69
Query 103 IVLINQHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFGGELPS 159
IVLINQHVAD+IRY+VDLH++++PTILEIPSKD+PYDPSKDSVMQR+KFFFGGELP+
Sbjct 70 IVLINQHVADDIRYMVDLHTKIIPTILEIPSKDKPYDPSKDSVMQRIKFFFGGELPT 126
> pfa:PF11_0412 vacuolar ATP synthase subunit F, putative; K02151
V-type H+-transporting ATPase subunit F [EC:3.6.3.14]
Length=128
Score = 166 bits (420), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 69/119 (57%), Positives = 97/119 (81%), Gaps = 0/119 (0%)
Query 39 RPHGAAELKICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHR 98
R +LKI +IGDED+V GFL+AGIG RDG+GK NF +V++KT + +IE F +++ +
Sbjct 7 RLFNETDLKIYIIGDEDSVVGFLLAGIGFRDGLGKKNFFIVNSKTNKSEIEEVFKEYSSK 66
Query 99 SDVGIVLINQHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFGGEL 157
D G++LINQ +AD IRYLVDLH +++PT+LEIPSKD+P+DP+KDS++QRVK FFGG++
Sbjct 67 HDCGVILINQQIADEIRYLVDLHDKILPTVLEIPSKDKPFDPNKDSIIQRVKLFFGGDI 125
> cpv:cgd5_850 vacuolar ATP synthase subunit F ; K02151 V-type
H+-transporting ATPase subunit F [EC:3.6.3.14]
Length=125
Score = 164 bits (415), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 72/113 (63%), Positives = 91/113 (80%), Gaps = 0/113 (0%)
Query 46 LKICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHRSDVGIVL 105
LKI +IGDEDTVAGFL+ GIG RD GKTNFL+VD+KT + IE F DF + D GI++
Sbjct 12 LKIYIIGDEDTVAGFLLTGIGARDPQGKTNFLIVDSKTPQSQIEETFKDFISQQDCGILM 71
Query 106 INQHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFGGELP 158
INQ +A+ IRYLV+ H +++PTILE+PSKD+PYD +KDSVMQR+K F+GG LP
Sbjct 72 INQTIAEEIRYLVNTHDKIIPTILEVPSKDKPYDAAKDSVMQRIKLFYGGSLP 124
> tpv:TP02_0445 vacuolar ATP synthase subunit F; K02151 V-type
H+-transporting ATPase subunit F [EC:3.6.3.14]
Length=121
Score = 145 bits (365), Expect = 8e-35, Method: Compositional matrix adjust.
Identities = 64/108 (59%), Positives = 83/108 (76%), Gaps = 0/108 (0%)
Query 47 KICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHRSDVGIVLI 106
KI +IGDEDTV GFL+AG+G +D +G+TN+ +V K + IE F + R D GI++I
Sbjct 11 KIYIIGDEDTVVGFLLAGVGSKDVLGRTNYTIVTPKFTKAQIEEVFKLYVSREDCGIIII 70
Query 107 NQHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFG 154
NQH+A+ IR L+DLH + VPTILEIPSK+ PYDPSKDSVMQ++K FFG
Sbjct 71 NQHIAEKIRTLLDLHDKFVPTILEIPSKEEPYDPSKDSVMQKIKVFFG 118
> bbo:BBOV_III005090 17.m07454; vacuolar ATP synthase subunit
F; K02151 V-type H+-transporting ATPase subunit F [EC:3.6.3.14]
Length=120
Score = 144 bits (362), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 61/116 (52%), Positives = 86/116 (74%), Gaps = 0/116 (0%)
Query 39 RPHGAAELKICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHR 98
R + E K+ +IGDEDTV GFLMAGIG +DG+G+TN+ ++ K +Q+IE F + +
Sbjct 3 RINELTEAKVYIIGDEDTVVGFLMAGIGSKDGLGQTNYKIITPKVSKQEIEDTFKQYVQK 62
Query 99 SDVGIVLINQHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFG 154
D GI++INQH+A+ I+ +DLHS +P ILEIPSK++PYDP+KDSV Q++K FG
Sbjct 63 KDCGIIIINQHIAEKIKTAIDLHSGPIPAILEIPSKEQPYDPNKDSVTQKIKVLFG 118
> ath:AT4G02620 vacuolar ATPase subunit F family protein; K02151
V-type H+-transporting ATPase subunit F [EC:3.6.3.14]
Length=128
Score = 136 bits (342), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 61/109 (55%), Positives = 83/109 (76%), Gaps = 0/109 (0%)
Query 48 ICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHRSDVGIVLIN 107
I +I DEDTV GFLMAG+G D KTN+L+VD+KT + IE AF +F+ R D+ I+L++
Sbjct 15 IAMIADEDTVVGFLMAGVGNVDIRRKTNYLIVDSKTTVRQIEDAFKEFSARDDIAIILLS 74
Query 108 QHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFGGE 156
Q++A+ IR+LVD +++ VP ILEIPSKD PYDP+ DSV+ RVK+ F E
Sbjct 75 QYIANMIRFLVDSYNKPVPAILEIPSKDHPYDPAHDSVLSRVKYLFSAE 123
> cel:ZK970.4 vha-9; Vacuolar H ATPase family member (vha-9);
K02151 V-type H+-transporting ATPase subunit F [EC:3.6.3.14]
Length=121
Score = 131 bits (329), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 62/115 (53%), Positives = 83/115 (72%), Gaps = 1/115 (0%)
Query 43 AAELKI-CVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHRSDV 101
AA+ KI VIGDEDTV GFL+ G+G + K N+L+VD +T Q+IE AF+ F R D+
Sbjct 4 AAKGKILAVIGDEDTVVGFLLGGVGELNKARKPNYLIVDKQTTVQEIEEAFNGFCARDDI 63
Query 102 GIVLINQHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFGGE 156
I+LINQH+A+ IRY VD H++ +P +LEIPSK+ PYDPSKDS++ R + F E
Sbjct 64 AIILINQHIAEMIRYAVDNHTQSIPAVLEIPSKEAPYDPSKDSILNRARGLFNPE 118
> dre:436799 atp6v1f, zgc:92923; ATPase, H+ transporting, V1 subunit
F (EC:3.6.3.14); K02151 V-type H+-transporting ATPase
subunit F [EC:3.6.3.14]
Length=119
Score = 121 bits (303), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 56/109 (51%), Positives = 75/109 (68%), Gaps = 0/109 (0%)
Query 48 ICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHRSDVGIVLIN 107
I VIGDEDT GFL+ GIG + K NFLVV+ +T +IE F F R+D+GI+LIN
Sbjct 8 IAVIGDEDTCTGFLLGGIGELNKNRKPNFLVVEKETSVTEIEETFKSFLARNDIGIILIN 67
Query 108 QHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFGGE 156
Q +A+ IR+ +D H + +P +LEIPSK+ PYD SKDS+++R K F E
Sbjct 68 QFIAEMIRHAIDAHMQSIPAVLEIPSKEHPYDASKDSILRRAKGMFSAE 116
> xla:398852 atp6v1f-b, MGC115045, MGC68786, Vma7, atp6s14, atp6v1f,
vatf; ATPase, H+ transporting, lysosomal 14kDa, V1 subunit
F; K02151 V-type H+-transporting ATPase subunit F [EC:3.6.3.14]
Length=122
Score = 120 bits (302), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 56/109 (51%), Positives = 77/109 (70%), Gaps = 0/109 (0%)
Query 48 ICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHRSDVGIVLIN 107
I VIGDEDTV GFL+ GIG + K NFLVV+ +T +IE F F +R D+GI+LIN
Sbjct 11 IAVIGDEDTVTGFLLGGIGELNKNRKPNFLVVEKETSVTEIEETFRSFLNRDDIGIILIN 70
Query 108 QHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFGGE 156
Q +A+ IR+++D H+ +P +LEIPSK+ PYD +KDS+++R K F E
Sbjct 71 QFIAEMIRHVIDTHTISIPAVLEIPSKEHPYDATKDSILRRAKGMFTME 119
> mmu:66144 Atp6v1f, 1110004G16Rik; ATPase, H+ transporting, lysosomal
V1 subunit F (EC:3.6.3.14); K02151 V-type H+-transporting
ATPase subunit F [EC:3.6.3.14]
Length=119
Score = 119 bits (297), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 55/109 (50%), Positives = 75/109 (68%), Gaps = 0/109 (0%)
Query 48 ICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHRSDVGIVLIN 107
I VIGDEDTV GFL+ GIG + NFLVV+ T +IE F F +R D+GI+LIN
Sbjct 8 IAVIGDEDTVTGFLLGGIGELNKNRHPNFLVVEKDTTINEIEDTFRQFLNRDDIGIILIN 67
Query 108 QHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFGGE 156
Q++A+ +R+ +D H R +P +LEIPSK+ PYD +KDS+++R K F E
Sbjct 68 QYIAEMVRHALDAHQRSIPAVLEIPSKEHPYDAAKDSILRRAKGMFTAE 116
> xla:403358 hypothetical protein MGC68592; K02151 V-type H+-transporting
ATPase subunit F [EC:3.6.3.14]
Length=122
Score = 119 bits (297), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 54/106 (50%), Positives = 75/106 (70%), Gaps = 0/106 (0%)
Query 48 ICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHRSDVGIVLIN 107
I +IGDEDTV GFL+ GIG + K NFLVV+ +T +IE F F +R D+GI+LIN
Sbjct 11 IAIIGDEDTVTGFLLGGIGELNKNRKPNFLVVEKETSVTEIEETFRSFLNRDDIGIILIN 70
Query 108 QHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFF 153
Q +A+ IR+ +D H+ +P +LEIPSK+ PYD +KDS+++R K F
Sbjct 71 QFIAEMIRHAIDAHTISIPAVLEIPSKEHPYDATKDSILRRAKGMF 116
> sce:YGR020C VMA7; Subunit F of the eight-subunit V1 peripheral
membrane domain of vacuolar H+-ATPase (V-ATPase), an electrogenic
proton pump found throughout the endomembrane system;
required for the V1 domain to assemble onto the vacuolar
membrane (EC:3.6.3.14); K02151 V-type H+-transporting ATPase
subunit F [EC:3.6.3.14]
Length=118
Score = 109 bits (273), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 57/110 (51%), Positives = 73/110 (66%), Gaps = 3/110 (2%)
Query 48 ICVIGDEDTVAGFLMAGIG-MRDGMGKTNFLVV-DTKTKRQDIERAFHDFTH-RSDVGIV 104
I VI DEDT G L+AGIG + + NF V + KT +++I F+ FT R D+ I+
Sbjct 8 IAVIADEDTTTGLLLAGIGQITPETQEKNFFVYQEGKTTKEEITDKFNHFTEERDDIAIL 67
Query 105 LINQHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFG 154
LINQH+A+NIR VD + P ILEIPSKD PYDP KDSV++RV+ FG
Sbjct 68 LINQHIAENIRARVDSFTNAFPAILEIPSKDHPYDPEKDSVLKRVRKLFG 117
> hsa:100505587 v-type proton ATPase subunit F-like
Length=116
Score = 104 bits (260), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 49/109 (44%), Positives = 71/109 (65%), Gaps = 0/109 (0%)
Query 48 ICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHRSDVGIVLIN 107
I VI D+DTV GFL+ IG + NFLVV+ T +IE F F +R D GI+LIN
Sbjct 8 IAVIRDKDTVTGFLLGSIGELNKNCHPNFLVVEKDTTINEIEDTFRQFLNRDDTGIILIN 67
Query 108 QHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFGGE 156
Q++A+ +++ +D H +PT+LEIPSK+ PY+ +KDS ++R + F E
Sbjct 68 QYIAEMVQHALDTHQHSIPTVLEIPSKEHPYEDAKDSTLRRARGMFTAE 116
> hsa:9296 ATP6V1F, ATP6S14, MGC117321, MGC126037, MGC126038,
VATF, Vma7; ATPase, H+ transporting, lysosomal 14kDa, V1 subunit
F (EC:3.6.3.14); K02151 V-type H+-transporting ATPase subunit
F [EC:3.6.3.14]
Length=147
Score = 101 bits (252), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 53/137 (38%), Positives = 75/137 (54%), Gaps = 28/137 (20%)
Query 48 ICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHD------------- 94
I VIGDEDTV GFL+ GIG + NFLVV+ T +IE F
Sbjct 8 IAVIGDEDTVTGFLLGGIGELNKNRHPNFLVVEKDTTINEIEDTFRSLGSLPGSVVEANP 67
Query 95 ---------------FTHRSDVGIVLINQHVADNIRYLVDLHSRVVPTILEIPSKDRPYD 139
F +R D+GI+LINQ++A+ +R+ +D H + +P +LEIPSK+ PYD
Sbjct 68 NQRDPPLWDEIDSRQFLNRDDIGIILINQYIAEMVRHALDAHQQSIPAVLEIPSKEHPYD 127
Query 140 PSKDSVMQRVKFFFGGE 156
+KDS+++R + F E
Sbjct 128 AAKDSILRRARGMFTAE 144
> dre:797834 cb606, sb:cb606; si:dkeyp-110c7.1
Length=394
Score = 30.0 bits (66), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 32/110 (29%), Positives = 45/110 (40%), Gaps = 17/110 (15%)
Query 42 GAAELKICVIGD---EDTVAGFLMAGIG---MRDGMGKTNFLVVDTKTKRQDIERAFHDF 95
G E IGD ED V G L AGIG + + G V +T+ RQ E F+
Sbjct 96 GGGERLDSAIGDSINEDAVVGCLSAGIGTMILSEPAGTDRQAVTNTEEGRQRREELFNTL 155
Query 96 THRSDVGIVLINQHVADNIRYLVDLHSR--VVPTILEIPSKDRPYDPSKD 143
S+ G D + +L +H + VV +L+ D + P D
Sbjct 156 NFLSEDG---------DTVLHLALIHEQWGVVQCLLQNIGMDNTWIPYLD 196
> ath:AT4G39660 AGT2; AGT2 (ALANINE:GLYOXYLATE AMINOTRANSFERASE
2); alanine-glyoxylate transaminase/ catalytic/ pyridoxal
phosphate binding / transaminase; K00827 alanine-glyoxylate
transaminase / (R)-3-amino-2-methylpropionate-pyruvate transaminase
[EC:2.6.1.44 2.6.1.40]
Length=476
Score = 29.6 bits (65), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 35/75 (46%), Gaps = 8/75 (10%)
Query 84 KRQDIERAFHDFTHRSDVGIVLINQHVADNIRYLVDLHSRVVPTILEIPSKDRPYD-PSK 142
+RQ ++RA D HR + ++ + + +I + P I + RPY PS
Sbjct 4 QRQLLKRATSDIYHRRAISLLRTDFSTSPSI-------ADAPPHIPPFVHQPRPYKGPSA 56
Query 143 DSVMQRVKFFFGGEL 157
D V+Q+ K F G L
Sbjct 57 DEVLQKRKKFLGPSL 71
> tgo:TGME49_051850 serine/threonine protein phosphatase 5, putative
(EC:3.1.2.15 3.1.3.16); K01090 protein phosphatase [EC:3.1.3.16]
Length=1086
Score = 29.6 bits (65), Expect = 4.0, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 28/62 (45%), Gaps = 0/62 (0%)
Query 72 GKTNFLVVDTKTKRQDIERAFHDFTHRSDVGIVLINQHVADNIRYLVDLHSRVVPTILEI 131
G LV DT + D+ F+ F S + L N +AD RY V++ + L+
Sbjct 323 GSRLVLVGDTHGQLNDVLWIFYKFGPPSATNVYLFNGDIADRGRYAVEIFMMLFAFKLQC 382
Query 132 PS 133
PS
Sbjct 383 PS 384
> cel:VC5.3 npa-1; Nematode Polyprotein Allergen related family
member (npa-1)
Length=1440
Score = 28.9 bits (63), Expect = 7.5, Method: Composition-based stats.
Identities = 23/61 (37%), Positives = 31/61 (50%), Gaps = 10/61 (16%)
Query 47 KICVIGDEDTVAGFLMAGIGMR-DGMGKT---NFLVV------DTKTKRQDIERAFHDFT 96
KI + D+ AG L+ G+ R +G K L V D+K KR++IE AF DF
Sbjct 471 KIKTMKDDKVAAGALVKGVVDRQEGEVKAIAEKMLSVCGEVYKDSKRKRREIEAAFKDFV 530
Query 97 H 97
H
Sbjct 531 H 531
Lambda K H
0.322 0.137 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3647184800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40