bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4302_orf1
Length=132
Score E
Sequences producing significant alignments: (Bits) Value
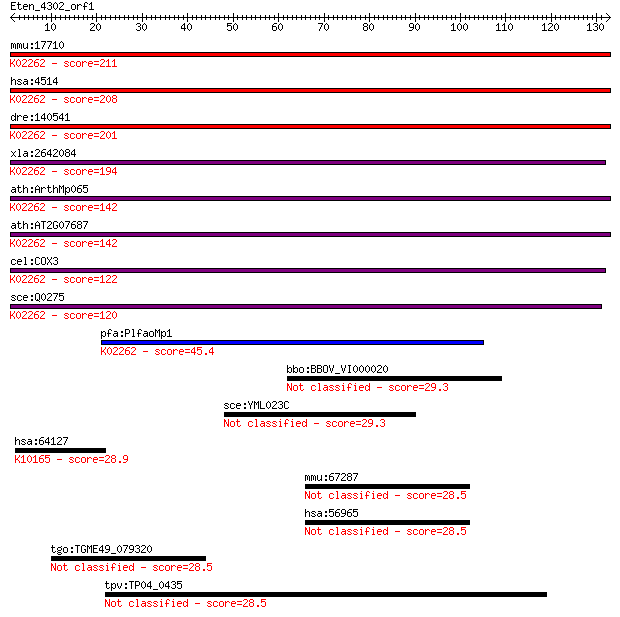
mmu:17710 COX3; cytochrome c oxidase subunit III (EC:1.9.3.1);... 211 6e-55
hsa:4514 COX3, COIII, MTCO3; cytochrome c oxidase III; K02262 ... 208 4e-54
dre:140541 COX3, mtco3; cytochrome c oxidase subunit III; K022... 201 4e-52
xla:2642084 COX3; cytochrome c oxidase subunit III; K02262 cyt... 194 4e-50
ath:ArthMp065 cox3; cytochrome c oxidase subunit 3; K02262 cyt... 142 2e-34
ath:AT2G07687 cytochrome c oxidase subunit 3 (EC:1.9.3.1); K02... 142 2e-34
cel:COX3 cytochrome c oxidase subunit III; K02262 cytochrome c... 122 2e-28
sce:Q0275 COX3, OXI2; Cox3p (EC:1.9.3.1); K02262 cytochrome c ... 120 2e-27
pfa:PlfaoMp1 coxIII; putative cytochrome oxidase III; K02262 c... 45.4 4e-05
bbo:BBOV_VI000020 cytochrome c oxidase subunit III 29.3 3.6
sce:YML023C NSE5; Essential subunit of the Mms21-Smc5-Smc6 com... 29.3 3.7
hsa:64127 NOD2, ACUG, BLAU, CARD15, CD, CLR16.3, IBD1, NLRC2, ... 28.9 4.3
mmu:67287 Parp6, 1700119G14Rik, 2310028P13Rik, 3110038K10Rik, ... 28.5 5.7
hsa:56965 PARP6, MGC131971, pART17; poly (ADP-ribose) polymera... 28.5 5.7
tgo:TGME49_079320 hypothetical protein 28.5 6.0
tpv:TP04_0435 hypothetical protein 28.5 6.1
> mmu:17710 COX3; cytochrome c oxidase subunit III (EC:1.9.3.1);
K02262 cytochrome c oxidase subunit III [EC:1.9.3.1]
Length=261
Score = 211 bits (536), Expect = 6e-55, Method: Compositional matrix adjust.
Identities = 103/132 (78%), Positives = 112/132 (84%), Gaps = 0/132 (0%)
Query 1 PTPELGGCWPPTGIHPLNPLEVPLLNTSVLLASGVSITGAPHSLIEGGRKHILQALFFPI 60
PT +LGGCWPPTGI PLNPLEVPLLNTSVLLASGVSIT A HSL+EG R H+ QAL I
Sbjct 108 PTHDLGGCWPPTGISPLNPLEVPLLNTSVLLASGVSITWAHHSLMEGKRNHMNQALLITI 167
Query 61 TLGVYFTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHVIIGSPFLIVCFFPQLKFHF 120
LG+YFT+LQASEY+E F+ISDG+YGSTFF+ATGFHGLHVIIGS FLIVC QLKFHF
Sbjct 168 MLGLYFTILQASEYFETSFSISDGIYGSTFFMATGFHGLHVIIGSTFLIVCLLRQLKFHF 227
Query 121 TSNPHFGFETAA 132
TS HFGFE AA
Sbjct 228 TSKHHFGFEAAA 239
> hsa:4514 COX3, COIII, MTCO3; cytochrome c oxidase III; K02262
cytochrome c oxidase subunit III [EC:1.9.3.1]
Length=261
Score = 208 bits (529), Expect = 4e-54, Method: Compositional matrix adjust.
Identities = 103/132 (78%), Positives = 112/132 (84%), Gaps = 0/132 (0%)
Query 1 PTPELGGCWPPTGIHPLNPLEVPLLNTSVLLASGVSITGAPHSLIEGGRKHILQALFFPI 60
PTP+LGG WPPTGI PLNPLEVPLLNTSVLLASGVSIT A HSL+E R ++QAL I
Sbjct 108 PTPQLGGHWPPTGITPLNPLEVPLLNTSVLLASGVSITWAHHSLMENNRNQMIQALLITI 167
Query 61 TLGVYFTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHVIIGSPFLIVCFFPQLKFHF 120
LG+YFTLLQASEY+E+PFTISDG+YGSTFFVATGFHGLHVIIGS FL +CF QL FHF
Sbjct 168 LLGLYFTLLQASEYFESPFTISDGIYGSTFFVATGFHGLHVIIGSTFLTICFIRQLMFHF 227
Query 121 TSNPHFGFETAA 132
TS HFGFE AA
Sbjct 228 TSKHHFGFEAAA 239
> dre:140541 COX3, mtco3; cytochrome c oxidase subunit III; K02262
cytochrome c oxidase subunit III [EC:1.9.3.1]
Length=261
Score = 201 bits (511), Expect = 4e-52, Method: Compositional matrix adjust.
Identities = 98/132 (74%), Positives = 110/132 (83%), Gaps = 0/132 (0%)
Query 1 PTPELGGCWPPTGIHPLNPLEVPLLNTSVLLASGVSITGAPHSLIEGGRKHILQALFFPI 60
PTPELGGCWPPTG+ L+P EVPLLNT+VLLASGV++T A HSL+EG RK +Q+L I
Sbjct 108 PTPELGGCWPPTGLTTLDPFEVPLLNTAVLLASGVTVTWAHHSLMEGERKQAIQSLALTI 167
Query 61 TLGVYFTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHVIIGSPFLIVCFFPQLKFHF 120
LG+YFT LQA EYYEAPFTI+DGVYGSTFFVATGFHGLHVIIGS FL VC Q+ FHF
Sbjct 168 LLGLYFTALQAMEYYEAPFTIADGVYGSTFFVATGFHGLHVIIGSTFLAVCLLRQVLFHF 227
Query 121 TSNPHFGFETAA 132
TS+ HFGFE AA
Sbjct 228 TSDHHFGFEAAA 239
> xla:2642084 COX3; cytochrome c oxidase subunit III; K02262 cytochrome
c oxidase subunit III [EC:1.9.3.1]
Length=260
Score = 194 bits (494), Expect = 4e-50, Method: Compositional matrix adjust.
Identities = 95/131 (72%), Positives = 107/131 (81%), Gaps = 0/131 (0%)
Query 1 PTPELGGCWPPTGIHPLNPLEVPLLNTSVLLASGVSITGAPHSLIEGGRKHILQALFFPI 60
PT ELG CWPPTGI PLNP EVPLLNT+VLLASGV++T A HS++ G RK +Q+L I
Sbjct 108 PTYELGECWPPTGITPLNPFEVPLLNTAVLLASGVTVTWAHHSIMHGDRKEAIQSLTLTI 167
Query 61 TLGVYFTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHVIIGSPFLIVCFFPQLKFHF 120
LG+YFT LQA EYYEAPFTI+DGVYGSTFFVATGFHGLHVIIGS FL VC Q+++HF
Sbjct 168 LLGLYFTALQAMEYYEAPFTIADGVYGSTFFVATGFHGLHVIIGSLFLSVCLLRQIQYHF 227
Query 121 TSNPHFGFETA 131
TS HFGFE A
Sbjct 228 TSKHHFGFEAA 238
> ath:ArthMp065 cox3; cytochrome c oxidase subunit 3; K02262 cytochrome
c oxidase subunit III [EC:1.9.3.1]
Length=265
Score = 142 bits (359), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 70/132 (53%), Positives = 88/132 (66%), Gaps = 0/132 (0%)
Query 1 PTPELGGCWPPTGIHPLNPLEVPLLNTSVLLASGVSITGAPHSLIEGGRKHILQALFFPI 60
P E+GG WPP GI L+P E+P LNT +L +SG ++T A H+++ G K + AL +
Sbjct 111 PAVEIGGIWPPKGIEVLDPWEIPFLNTPILPSSGAAVTWAHHAILAGKEKRAVYALVATV 170
Query 61 TLGVYFTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHVIIGSPFLIVCFFPQLKFHF 120
L + FT Q EYY+APFTISD +YGSTFF+ATGFHG HVIIG+ FLI+C Q H
Sbjct 171 LLALVFTGFQGMEYYQAPFTISDSIYGSTFFLATGFHGFHVIIGTLFLIICGIRQYLGHL 230
Query 121 TSNPHFGFETAA 132
T H GFE AA
Sbjct 231 TKEHHVGFEAAA 242
> ath:AT2G07687 cytochrome c oxidase subunit 3 (EC:1.9.3.1); K02262
cytochrome c oxidase subunit III [EC:1.9.3.1]
Length=265
Score = 142 bits (359), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 70/132 (53%), Positives = 88/132 (66%), Gaps = 0/132 (0%)
Query 1 PTPELGGCWPPTGIHPLNPLEVPLLNTSVLLASGVSITGAPHSLIEGGRKHILQALFFPI 60
P E+GG WPP GI L+P E+P LNT +L +SG ++T A H+++ G K + AL +
Sbjct 111 PAVEIGGIWPPKGIEVLDPWEIPFLNTPILPSSGAAVTWAHHAILAGKEKRAVYALVATV 170
Query 61 TLGVYFTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHVIIGSPFLIVCFFPQLKFHF 120
L + FT Q EYY+APFTISD +YGSTFF+ATGFHG HVIIG+ FLI+C Q H
Sbjct 171 LLALVFTGFQGMEYYQAPFTISDSIYGSTFFLATGFHGFHVIIGTLFLIICGIRQYLGHL 230
Query 121 TSNPHFGFETAA 132
T H GFE AA
Sbjct 231 TKEHHVGFEAAA 242
> cel:COX3 cytochrome c oxidase subunit III; K02262 cytochrome
c oxidase subunit III [EC:1.9.3.1]
Length=255
Score = 122 bits (307), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 64/131 (48%), Positives = 82/131 (62%), Gaps = 2/131 (1%)
Query 1 PTPELGGCWPPTGIHPLNPLEVPLLNTSVLLASGVSITGAPHSLIEGGRKHILQALFFPI 60
P ELG W P G+H +NP VPLLNT +LL+SGV++T A HSL+ K ++
Sbjct 104 PVHELGETWSPFGMHLVNPFGVPLLNTIILLSSGVTVTWAHHSLLS--NKSCTNSMILTC 161
Query 61 TLGVYFTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHVIIGSPFLIVCFFPQLKFHF 120
L YFT +Q EY EA F+I+DGV+GS F+++TGFHG+HV+ G FL F LK HF
Sbjct 162 LLAAYFTGIQLMEYMEASFSIADGVFGSIFYLSTGFHGIHVLCGGLFLAFNFLRLLKNHF 221
Query 121 TSNPHFGFETA 131
N H G E A
Sbjct 222 NYNHHLGLEFA 232
> sce:Q0275 COX3, OXI2; Cox3p (EC:1.9.3.1); K02262 cytochrome
c oxidase subunit III [EC:1.9.3.1]
Length=269
Score = 120 bits (300), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 59/130 (45%), Positives = 77/130 (59%), Gaps = 0/130 (0%)
Query 1 PTPELGGCWPPTGIHPLNPLEVPLLNTSVLLASGVSITGAPHSLIEGGRKHILQALFFPI 60
P LG CWPP GI + P E+PLLNT +LL+SG ++T + H+LI G R L L
Sbjct 116 PDVTLGACWPPVGIEAVQPTELPLLNTIILLSSGATVTYSHHALIAGNRNKALSGLLITF 175
Query 61 TLGVYFTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHVIIGSPFLIVCFFPQLKFHF 120
L V F Q EY A FTISDGVYGS F+ TG H LH+++ + L V ++ +H
Sbjct 176 WLIVIFVTCQYIEYTNAAFTISDGVYGSVFYAGTGLHFLHMVMLAAMLGVNYWRMRNYHL 235
Query 121 TSNPHFGFET 130
T+ H G+ET
Sbjct 236 TAGHHVGYET 245
> pfa:PlfaoMp1 coxIII; putative cytochrome oxidase III; K02262
cytochrome c oxidase subunit III [EC:1.9.3.1]
Length=279
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 42/85 (49%), Gaps = 1/85 (1%)
Query 21 EVPLLNTSVLLASGVSITGAPHSLIEGGRKHILQALFFPI-TLGVYFTLLQASEYYEAPF 79
+ +L + +LAS +T IE G + ++ I LG F LQ +EY +
Sbjct 134 RMLILTITFILASASCMTACLQVFIEKGMSFEISSIICIIYLLGECFASLQTTEYLHLSY 193
Query 80 TISDGVYGSTFFVATGFHGLHVIIG 104
I+D VY + F+ TG H HV+IG
Sbjct 194 HINDTVYTTLFYCVTGLHFSHVVIG 218
> bbo:BBOV_VI000020 cytochrome c oxidase subunit III
Length=228
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 22/47 (46%), Gaps = 0/47 (0%)
Query 62 LGVYFTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHVIIGSPFL 108
+G F Q EY ++D + F + TG H LHV +G F+
Sbjct 141 IGSLFLSYQGDEYTFLQCGLNDSWFSLAFLIITGLHSLHVCVGVLFI 187
> sce:YML023C NSE5; Essential subunit of the Mms21-Smc5-Smc6 complex;
required for cell viability and DNA repair
Length=556
Score = 29.3 bits (64), Expect = 3.7, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query 48 GRKHILQALFFPITLGVYFTLLQASEYYEAPFTISDGVYGST 89
G+ H L + F I L V FTL SEYY+ P ++ Y ++
Sbjct 56 GKSHNLAVIPFDIIL-VLFTLSTLSEYYKEPILRANDPYNTS 96
> hsa:64127 NOD2, ACUG, BLAU, CARD15, CD, CLR16.3, IBD1, NLRC2,
NOD2B, PSORAS1; nucleotide-binding oligomerization domain
containing 2; K10165 nucleotide-binding oligomerization domain-containing
protein 2
Length=1040
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 9/20 (45%), Positives = 13/20 (65%), Gaps = 0/20 (0%)
Query 2 TPELGGCWPPTGIHPLNPLE 21
+P+L GCW P +HP L+
Sbjct 116 SPKLHGCWDPHSLHPARDLQ 135
> mmu:67287 Parp6, 1700119G14Rik, 2310028P13Rik, 3110038K10Rik,
C030013N01Rik, PARP-6; poly (ADP-ribose) polymerase family,
member 6 (EC:2.4.2.30)
Length=630
Score = 28.5 bits (62), Expect = 5.7, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 19/36 (52%), Gaps = 4/36 (11%)
Query 66 FTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHV 101
F LL + EA F + +YGSTF FHG H+
Sbjct 446 FLLLSSPPAKEARFRTAKKLYGSTF----AFHGSHI 477
> hsa:56965 PARP6, MGC131971, pART17; poly (ADP-ribose) polymerase
family, member 6 (EC:2.4.2.30)
Length=630
Score = 28.5 bits (62), Expect = 5.7, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 19/36 (52%), Gaps = 4/36 (11%)
Query 66 FTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHV 101
F LL + EA F + +YGSTF FHG H+
Sbjct 446 FLLLSSPPAKEARFRTAKKLYGSTF----AFHGSHI 477
> tgo:TGME49_079320 hypothetical protein
Length=4967
Score = 28.5 bits (62), Expect = 6.0, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 24/38 (63%), Gaps = 4/38 (10%)
Query 10 PPTGIHPLNPL--EVPLLNTSVLLASGVSITG--APHS 43
PP+G+HP++P ++PLL + L S + G APH+
Sbjct 3222 PPSGLHPVSPSAPQLPLLVSPAQLGSEAAHAGQPAPHA 3259
> tpv:TP04_0435 hypothetical protein
Length=1804
Score = 28.5 bits (62), Expect = 6.1, Method: Composition-based stats.
Identities = 22/97 (22%), Positives = 40/97 (41%), Gaps = 12/97 (12%)
Query 22 VPLLNTSVLLASGVSITGAPHSLIEGGRKHILQALFFPITLGVYFTLLQASEYYEAPFTI 81
+P++ + LL + IT + IE + G+ + L E +
Sbjct 756 LPMITSKHLLNDSIEITRYKGTRIETCKHR-----------GIIMSYLSVMFINEMGVEL 804
Query 82 SDGVYGSTFFVATGFHGLHVIIGSPFLIVCFFPQLKF 118
D V+G + G+H ++ I P+ V F+P +KF
Sbjct 805 FDSVHGDGLSIIKGYHDIYYNI-PPYRHVPFWPNVKF 840
Lambda K H
0.326 0.145 0.468
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2099897216
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40