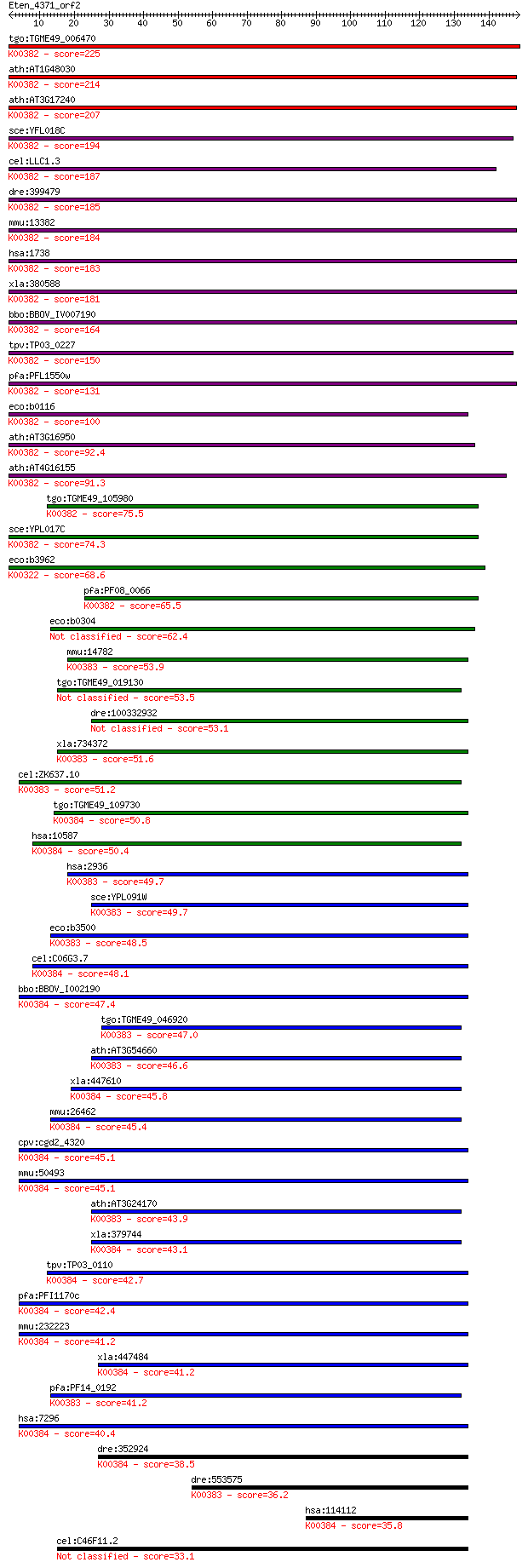
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4371_orf2
Length=148
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_006470 dihydrolipoyl dehydrogenase, putative (EC:1.... 225 5e-59
ath:AT1G48030 mtLPD1; mtLPD1 (mitochondrial lipoamide dehydrog... 214 1e-55
ath:AT3G17240 mtLPD2; mtLPD2 (LIPOAMIDE DEHYDROGENASE 2); ATP ... 207 1e-53
sce:YFL018C LPD1, HPD1; Dihydrolipoamide dehydrogenase, the li... 194 1e-49
cel:LLC1.3 hypothetical protein; K00382 dihydrolipoamide dehyd... 187 1e-47
dre:399479 dldh, wu:fb24b05; dihydrolipoamide dehydrogenase (E... 185 4e-47
mmu:13382 Dld, AI315664, AI746344; dihydrolipoamide dehydrogen... 184 8e-47
hsa:1738 DLD, DLDH, E3, GCSL, LAD, PHE3; dihydrolipoamide dehy... 183 2e-46
xla:380588 dld, MGC68940; dihydrolipoamide dehydrogenase (EC:1... 181 5e-46
bbo:BBOV_IV007190 23.m05858; dihydrolipoamide dehydrogenase (E... 164 1e-40
tpv:TP03_0227 dihydrolipoamide dehydrogenase (EC:1.8.1.4); K00... 150 2e-36
pfa:PFL1550w lipoamide dehydrogenase (EC:1.8.1.4); K00382 dihy... 131 8e-31
eco:b0116 lpd, dhl, ECK0115, JW0112, lpdA; lipoamide dehydroge... 100 2e-21
ath:AT3G16950 LPD1; LPD1 (LIPOAMIDE DEHYDROGENASE 1); dihydrol... 92.4 5e-19
ath:AT4G16155 dihydrolipoyl dehydrogenase; K00382 dihydrolipoa... 91.3 1e-18
tgo:TGME49_105980 dihydrolipoyl dehydrogenase protein, putativ... 75.5 5e-14
sce:YPL017C IRC15; Irc15p; K00382 dihydrolipoamide dehydrogena... 74.3 1e-13
eco:b3962 sthA, ECK3954, JW5551, sth, udhA; pyridine nucleotid... 68.6 7e-12
pfa:PF08_0066 lipoamide dehydrogenase, putative (EC:1.8.1.4); ... 65.5 5e-11
eco:b0304 ykgC, ECK0303, JW5040; predicted pyridine nucleotide... 62.4 6e-10
mmu:14782 Gsr, AI325518, D8Ertd238e, Gr-1, Gr1; glutathione re... 53.9 2e-07
tgo:TGME49_019130 glutathione reductase, putative (EC:1.8.1.7) 53.5 2e-07
dre:100332932 glutathione reductase-like 53.1 3e-07
xla:734372 gsr, MGC84926; glutathione reductase (EC:1.8.1.7); ... 51.6 8e-07
cel:ZK637.10 trxr-2; ThioRedoXin Reductase family member (trxr... 51.2 1e-06
tgo:TGME49_109730 thioredoxin reductase, putative (EC:1.8.1.7)... 50.8 1e-06
hsa:10587 TXNRD2, SELZ, TR, TR-BETA, TR3, TRXR2; thioredoxin r... 50.4 2e-06
hsa:2936 GSR, MGC78522; glutathione reductase (EC:1.8.1.7); K0... 49.7 3e-06
sce:YPL091W GLR1, LPG17; Cytosolic and mitochondrial glutathio... 49.7 4e-06
eco:b3500 gor, ECK3485, gorA, JW3467; glutathione oxidoreducta... 48.5 7e-06
cel:C06G3.7 trxr-1; ThioRedoXin Reductase family member (trxr-... 48.1 9e-06
bbo:BBOV_I002190 19.m02214; thiodoxin reductase (EC:1.8.1.9); ... 47.4 2e-05
tgo:TGME49_046920 glutathione reductase, putative (EC:1.8.1.7)... 47.0 2e-05
ath:AT3G54660 GR; GR (GLUTATHIONE REDUCTASE); ATP binding / gl... 46.6 3e-05
xla:447610 txnrd1, MGC85342; thioredoxin reductase 1 (EC:1.8.1... 45.8 5e-05
mmu:26462 Txnrd2, AA118373, ESTM573010, TGR, Tr3, Trxr2, Trxrd... 45.4 7e-05
cpv:cgd2_4320 thioredoxin reductase 1 ; K00384 thioredoxin red... 45.1 8e-05
mmu:50493 Txnrd1, TR, TR1, TrxR1; thioredoxin reductase 1 (EC:... 45.1 9e-05
ath:AT3G24170 ATGR1; ATGR1 (glutathione-disulfide reductase); ... 43.9 2e-04
xla:379744 txnrd2, MGC69182; thioredoxin reductase 2 (EC:1.8.1... 43.1 3e-04
tpv:TP03_0110 thioredoxin reductase (EC:1.6.4.5); K00384 thior... 42.7 4e-04
pfa:PFI1170c thioredoxin reductase (EC:1.8.1.9); K00384 thiore... 42.4 6e-04
mmu:232223 Txnrd3, AI196535, TR2, Tgr; thioredoxin reductase 3... 41.2 0.001
xla:447484 txnrd3, MGC81848; thioredoxin reductase 3 (EC:1.8.1... 41.2 0.001
pfa:PF14_0192 glutathione reductase (EC:1.8.1.7); K00383 gluta... 41.2 0.001
hsa:7296 TXNRD1, GRIM-12, MGC9145, TR, TR1, TRXR1, TXNR; thior... 40.4 0.002
dre:352924 txnrd1, TrxR1, cb682, fb83a08, wu:fb83a08; thioredo... 38.5 0.007
dre:553575 MGC110010; zgc:110010 (EC:1.8.1.7); K00383 glutathi... 36.2 0.042
hsa:114112 TXNRD3, TGR, TR2, TRXR3; thioredoxin reductase 3 (E... 35.8 0.046
cel:C46F11.2 hypothetical protein 33.1 0.31
> tgo:TGME49_006470 dihydrolipoyl dehydrogenase, putative (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=519
Score = 225 bits (573), Expect = 5e-59, Method: Compositional matrix adjust.
Identities = 105/148 (70%), Positives = 125/148 (84%), Gaps = 0/148 (0%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAF 60
AHKAEEEGIACVE +AG G+G V ++TIP+VIYTHPE+AGVGKTEE+LKA GV Y +G F
Sbjct 372 AHKAEEEGIACVEMIAGVGEGHVNYETIPSVIYTHPEIAGVGKTEEELKANGVSYNKGTF 431
Query 61 PFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLG 120
PFAANSRARA + G VKVL K++DK+LG WI+GP AGELI + VL MEYGA++EDLG
Sbjct 432 PFAANSRARANDVATGFVKVLAHKDSDKLLGAWIMGPEAGELIGQLVLGMEYGAAAEDLG 491
Query 121 RVCHAHPTQSEALKEACMACFDKPIHIA 148
R C +HPT SEA+KEACMAC+DKPIH+A
Sbjct 492 RTCVSHPTLSEAVKEACMACYDKPIHMA 519
> ath:AT1G48030 mtLPD1; mtLPD1 (mitochondrial lipoamide dehydrogenase
1); ATP binding / dihydrolipoyl dehydrogenase; K00382
dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=507
Score = 214 bits (544), Expect = 1e-55, Method: Compositional matrix adjust.
Identities = 104/147 (70%), Positives = 121/147 (82%), Gaps = 1/147 (0%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAF 60
AHKAEE+G+ACVE +AGK G V +D +P V+YTHPE+A VGKTEEQLK EGV Y+ G F
Sbjct 362 AHKAEEDGVACVEFIAGK-HGHVDYDKVPGVVYTHPEVASVGKTEEQLKKEGVSYRVGKF 420
Query 61 PFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLG 120
PF ANSRA+A +++GLVK+L KETDKILGV I+ PNAGELI EAVLA+ Y ASSED+
Sbjct 421 PFMANSRAKAIDNAEGLVKILADKETDKILGVHIMAPNAGELIHEAVLAINYDASSEDIA 480
Query 121 RVCHAHPTQSEALKEACMACFDKPIHI 147
RVCHAHPT SEALKEA MA +DKPIHI
Sbjct 481 RVCHAHPTMSEALKEAAMATYDKPIHI 507
> ath:AT3G17240 mtLPD2; mtLPD2 (LIPOAMIDE DEHYDROGENASE 2); ATP
binding / dihydrolipoyl dehydrogenase; K00382 dihydrolipoamide
dehydrogenase [EC:1.8.1.4]
Length=507
Score = 207 bits (526), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 100/147 (68%), Positives = 119/147 (80%), Gaps = 1/147 (0%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAF 60
AHKAEE+G+ACVE +AGK G V +D +P V+YT+PE+A VGKTEEQLK EGV Y G F
Sbjct 362 AHKAEEDGVACVEFIAGK-HGHVDYDKVPGVVYTYPEVASVGKTEEQLKKEGVSYNVGKF 420
Query 61 PFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLG 120
PF ANSRA+A ++G+VK+L KETDKILGV I+ PNAGELI EAVLA+ Y ASSED+
Sbjct 421 PFMANSRAKAIDTAEGMVKILADKETDKILGVHIMSPNAGELIHEAVLAINYDASSEDIA 480
Query 121 RVCHAHPTQSEALKEACMACFDKPIHI 147
RVCHAHPT SEA+KEA MA +DKPIH+
Sbjct 481 RVCHAHPTMSEAIKEAAMATYDKPIHM 507
> sce:YFL018C LPD1, HPD1; Dihydrolipoamide dehydrogenase, the
lipoamide dehydrogenase component (E3) of the pyruvate dehydrogenase
and 2-oxoglutarate dehydrogenase multi-enzyme complexes
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=499
Score = 194 bits (492), Expect = 1e-49, Method: Compositional matrix adjust.
Identities = 95/146 (65%), Positives = 115/146 (78%), Gaps = 1/146 (0%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAF 60
AHKAEEEGIA VE L G G V ++ IP+V+Y+HPE+A VGKTEEQLK G+ YK G F
Sbjct 354 AHKAEEEGIAAVEMLK-TGHGHVNYNNIPSVMYSHPEVAWVGKTEEQLKEAGIDYKIGKF 412
Query 61 PFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLG 120
PFAANSRA+ D++G VK+L +T++ILG I+GPNAGE+IAEA LA+EYGAS+ED+
Sbjct 413 PFAANSRAKTNQDTEGFVKILIDSKTERILGAHIIGPNAGEMIAEAGLALEYGASAEDVA 472
Query 121 RVCHAHPTQSEALKEACMACFDKPIH 146
RVCHAHPT SEA KEA MA +DK IH
Sbjct 473 RVCHAHPTLSEAFKEANMAAYDKAIH 498
> cel:LLC1.3 hypothetical protein; K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=495
Score = 187 bits (474), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 90/141 (63%), Positives = 111/141 (78%), Gaps = 1/141 (0%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAF 60
AHKAE+EGI CVE +AG G + ++ +P+V+YTHPE+A VGK EEQLK EGV YK G F
Sbjct 348 AHKAEDEGILCVEGIAG-GPVHIDYNCVPSVVYTHPEVAWVGKAEEQLKQEGVAYKIGKF 406
Query 61 PFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLG 120
PF ANSRA+ D +G VKVL K+TD++LGV I+GPNAGE+IAEA LAMEYGAS+ED+
Sbjct 407 PFVANSRAKTNNDQEGFVKVLADKQTDRMLGVHIIGPNAGEMIAEATLAMEYGASAEDVA 466
Query 121 RVCHAHPTQSEALKEACMACF 141
RVCH HPT SEA +EA +A +
Sbjct 467 RVCHPHPTLSEAFREANLAAY 487
> dre:399479 dldh, wu:fb24b05; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=507
Score = 185 bits (470), Expect = 4e-47, Method: Compositional matrix adjust.
Identities = 95/148 (64%), Positives = 115/148 (77%), Gaps = 2/148 (1%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAF 60
AHKAE+EGI CVE +AG G + ++ +P+VIYTHPE+A VGKTEEQLK EGV YK G F
Sbjct 361 AHKAEDEGIICVEGMAG-GAVHIDYNCVPSVIYTHPEVAWVGKTEEQLKEEGVPYKVGKF 419
Query 61 PFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLG 120
PFAANSRA+ D+DGLVK+L+ K+TD++LG ILG AGE+I EA LAMEYGAS ED+
Sbjct 420 PFAANSRAKTNADTDGLVKILSHKDTDRMLGAHILGSGAGEMINEAALAMEYGASCEDVA 479
Query 121 RVCHAHPTQSEALKEACM-ACFDKPIHI 147
RVCHAHPT SEA +EA + A F K I+
Sbjct 480 RVCHAHPTVSEAFREANLAASFGKAINF 507
> mmu:13382 Dld, AI315664, AI746344; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=509
Score = 184 bits (468), Expect = 8e-47, Method: Compositional matrix adjust.
Identities = 91/148 (61%), Positives = 115/148 (77%), Gaps = 2/148 (1%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAF 60
AHKAE+EGI CVE +AG G + ++ +P+VIYTHPE+A VGK+EEQLK EG+ +K G F
Sbjct 363 AHKAEDEGIICVEGMAG-GAVHIDYNCVPSVIYTHPEVAWVGKSEEQLKEEGIEFKIGKF 421
Query 61 PFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLG 120
PFAANSRA+ D+DG+VK+L K TD++LG ILGP AGE++ EA LA+EYGAS ED+
Sbjct 422 PFAANSRAKTNADTDGMVKILGHKSTDRVLGAHILGPGAGEMVNEAALALEYGASCEDIA 481
Query 121 RVCHAHPTQSEALKEACM-ACFDKPIHI 147
RVCHAHPT SEA +EA + A F KPI+
Sbjct 482 RVCHAHPTLSEAFREANLAAAFGKPINF 509
> hsa:1738 DLD, DLDH, E3, GCSL, LAD, PHE3; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=509
Score = 183 bits (464), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 91/148 (61%), Positives = 114/148 (77%), Gaps = 2/148 (1%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAF 60
AHKAE+EGI CVE +AG G + ++ +P+VIYTHPE+A VGK+EEQLK EG+ YK G F
Sbjct 363 AHKAEDEGIICVEGMAG-GAVHIDYNCVPSVIYTHPEVAWVGKSEEQLKEEGIEYKVGKF 421
Query 61 PFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLG 120
PFAANSRA+ D+DG+VK+L K TD++LG ILGP AGE++ EA LA+EYGAS ED+
Sbjct 422 PFAANSRAKTNADTDGMVKILGQKSTDRVLGAHILGPGAGEMVNEAALALEYGASCEDIA 481
Query 121 RVCHAHPTQSEALKEACM-ACFDKPIHI 147
RVCHAHPT SEA +EA + A F K I+
Sbjct 482 RVCHAHPTLSEAFREANLAASFGKSINF 509
> xla:380588 dld, MGC68940; dihydrolipoamide dehydrogenase (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=509
Score = 181 bits (460), Expect = 5e-46, Method: Compositional matrix adjust.
Identities = 93/148 (62%), Positives = 114/148 (77%), Gaps = 2/148 (1%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAF 60
AHKAE+EGI CVE +AG G + ++ +P+VIYTHPE+A VGK+EEQLK EG YK G F
Sbjct 363 AHKAEDEGIICVEGMAG-GAVHIDYNCVPSVIYTHPEVAWVGKSEEQLKEEGTEYKVGKF 421
Query 61 PFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLG 120
PFAANSRA+ D+DGLVK+L+ K TD++LG ILG +AGE+I EA LAMEYGAS ED+
Sbjct 422 PFAANSRAKTNADTDGLVKILSHKTTDRMLGAHILGASAGEMINEAALAMEYGASCEDVA 481
Query 121 RVCHAHPTQSEALKEACM-ACFDKPIHI 147
RVCHAHPT SEA +EA + A F K I+
Sbjct 482 RVCHAHPTVSEAFREANLAASFGKAINF 509
> bbo:BBOV_IV007190 23.m05858; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=481
Score = 164 bits (415), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 81/147 (55%), Positives = 103/147 (70%), Gaps = 1/147 (0%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAF 60
AHKAEE+G + + GK G + D IP VIYTHPE+AG+GK+E+ LK+ G+ YK+ F
Sbjct 336 AHKAEEDGSIALGHILGKDLGHINWDHIPMVIYTHPEVAGIGKSEQVLKSNGIEYKKATF 395
Query 61 PFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLG 120
PFAANSRAR GD DG VK+L K+ +KILG WI+GP+A ELI + + M G ++ D+
Sbjct 396 PFAANSRARIAGDVDGFVKILADKD-NKILGGWIVGPHASELIGQITIMMACGLTTVDVA 454
Query 121 RVCHAHPTQSEALKEACMACFDKPIHI 147
+VC AHPT SEALKEACMA K H
Sbjct 455 KVCFAHPTVSEALKEACMAVHHKATHF 481
> tpv:TP03_0227 dihydrolipoamide dehydrogenase (EC:1.8.1.4); K00382
dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=499
Score = 150 bits (378), Expect = 2e-36, Method: Composition-based stats.
Identities = 74/157 (47%), Positives = 105/157 (66%), Gaps = 12/157 (7%)
Query 1 AHKAEEEGIACVERLAGK-----------GDGTVRHDTIPAVIYTHPELAGVGKTEEQLK 49
AHKAEE+G+ + + GK G V + IP+VIYT PE+AGVG+TE+ L+
Sbjct 343 AHKAEEDGLIALGHILGKSFVHHPQGVTLGSVQVVPNVIPSVIYTEPEIAGVGETEQNLQ 402
Query 50 AEGVVYKRGAFPFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLA 109
GV YK+ FPF ANSRA+ +SDG +K+L+ +E +K+LG W++GP+ E+I LA
Sbjct 403 KLGVKYKKSVFPFMANSRAKIYNESDGFIKLLSTEE-NKLLGAWMIGPHVSEMIHTTALA 461
Query 110 MEYGASSEDLGRVCHAHPTQSEALKEACMACFDKPIH 146
+ YGASSED+ R+C AHP+ SEA+KE+ + KP+H
Sbjct 462 ITYGASSEDVTRMCFAHPSLSEAIKESSLGIHFKPLH 498
> pfa:PFL1550w lipoamide dehydrogenase (EC:1.8.1.4); K00382 dihydrolipoamide
dehydrogenase [EC:1.8.1.4]
Length=512
Score = 131 bits (329), Expect = 8e-31, Method: Composition-based stats.
Identities = 67/153 (43%), Positives = 94/153 (61%), Gaps = 6/153 (3%)
Query 1 AHKAEEEGIACVERL------AGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVV 54
AHKAEEEG L K + +D +P+VIYTHPE+A VG E + K +
Sbjct 360 AHKAEEEGYLLANILFDELKNNKKKKAHINYDLVPSVIYTHPEVATVGYNEAKCKELNMN 419
Query 55 YKRGAFPFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGA 114
+K +FPFAANSR+R D DGL+K++ K+T++ILG I+G NA +LI + +
Sbjct 420 FKSVSFPFAANSRSRTIDDYDGLIKLIVEKDTNRILGSQIIGNNASDLILPLSIYVANNG 479
Query 115 SSEDLGRVCHAHPTQSEALKEACMACFDKPIHI 147
SS+ L ++ +AHPT SE +KE + FDKPIH+
Sbjct 480 SSKSLSKIIYAHPTFSEVIKEVALQSFDKPIHM 512
> eco:b0116 lpd, dhl, ECK0115, JW0112, lpdA; lipoamide dehydrogenase,
E3 component is part of three enzyme complexes (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=474
Score = 100 bits (248), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 56/137 (40%), Positives = 84/137 (61%), Gaps = 9/137 (6%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRH----DTIPAVIYTHPELAGVGKTEEQLKAEGVVYK 56
AHK EG E +AGK +H IP++ YT PE+A VG TE++ K +G+ Y+
Sbjct 321 AHKGVHEGHVAAEVIAGK-----KHYFDPKVIPSIAYTEPEVAWVGLTEKEAKEKGISYE 375
Query 57 RGAFPFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASS 116
FP+AA+ RA A+ +DG+ K++ KE+ +++G I+G N GEL+ E LA+E G +
Sbjct 376 TATFPWAASGRAIASDCADGMTKLIFDKESHRVIGGAIVGTNGGELLGEIGLAIEMGCDA 435
Query 117 EDLGRVCHAHPTQSEAL 133
ED+ HAHPT E++
Sbjct 436 EDIALTIHAHPTLHESV 452
> ath:AT3G16950 LPD1; LPD1 (LIPOAMIDE DEHYDROGENASE 1); dihydrolipoyl
dehydrogenase; K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=623
Score = 92.4 bits (228), Expect = 5e-19, Method: Composition-based stats.
Identities = 55/139 (39%), Positives = 80/139 (57%), Gaps = 5/139 (3%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLK----AEGVVYK 56
AH A +GI+ VE+++G+ D + H +IPA +THPE++ VG TE Q K EG
Sbjct 411 AHAASAQGISVVEQVSGR-DHVLNHLSIPAACFTHPEISMVGLTEPQAKEKGEKEGFKVS 469
Query 57 RGAFPFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASS 116
F AN++A A + +G+ K++ + +ILGV I G +A +LI EA A+ G
Sbjct 470 VVKTSFKANTKALAENEGEGIAKMIYRPDNGEILGVHIFGLHAADLIHEASNAIALGTRI 529
Query 117 EDLGRVCHAHPTQSEALKE 135
+D+ HAHPT SE L E
Sbjct 530 QDIKLAVHAHPTLSEVLDE 548
> ath:AT4G16155 dihydrolipoyl dehydrogenase; K00382 dihydrolipoamide
dehydrogenase [EC:1.8.1.4]
Length=630
Score = 91.3 bits (225), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 57/149 (38%), Positives = 82/149 (55%), Gaps = 6/149 (4%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLK----AEGVVYK 56
AH A +GI+ VE++ G+ D + H +IPA +THPE++ VG TE Q + EG
Sbjct 471 AHAASAQGISVVEQVTGR-DHVLNHLSIPAACFTHPEISMVGLTEPQAREKAEKEGFKVS 529
Query 57 RGAFPFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASS 116
F AN++A A + +GL K++ + +ILGV I G +A +LI EA A+ G
Sbjct 530 IAKTSFKANTKALAENEGEGLAKMIYRPDNGEILGVHIFGLHAADLIHEASNAIALGTRI 589
Query 117 EDLGRVCHAHPTQSEALKEACMAC-FDKP 144
+D+ HAHPT SE + E A D P
Sbjct 590 QDIKLAVHAHPTLSEVVDELFKAAKVDSP 618
> tgo:TGME49_105980 dihydrolipoyl dehydrogenase protein, putative
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=636
Score = 75.5 bits (184), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 52/129 (40%), Positives = 74/129 (57%), Gaps = 5/129 (3%)
Query 12 VERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEE---QLKA-EGVVYKRGAFPFAANSR 67
VE +AG+ TV IPA +T PE+A +G TEE +L A +G + F AN++
Sbjct 499 VETIAGRPR-TVNVKHIPAACFTSPEIAFIGDTEEAAMELGAKDGFEVGKSVSHFRANTK 557
Query 68 ARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHP 127
A A G+ +G++KVL K+T KILG ++G +A +LI E A+ S +DL H HP
Sbjct 558 AIAEGEGEGILKVLYRKDTGKILGCHMIGIHASDLIQECATAITNDISVKDLAFTVHTHP 617
Query 128 TQSEALKEA 136
T SE + A
Sbjct 618 TLSEVVDAA 626
> sce:YPL017C IRC15; Irc15p; K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=499
Score = 74.3 bits (181), Expect = 1e-13, Method: Composition-based stats.
Identities = 47/143 (32%), Positives = 74/143 (51%), Gaps = 7/143 (4%)
Query 1 AHKAEEEGIACVERLAGKG-DGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGA 59
A KAEE+ I ++ + G DGT P V+Y P++ VG TEE L + Y++G
Sbjct 340 ALKAEEQAIRAIQSIGCTGSDGTSNCGFPPNVLYCQPQIGWVGYTEEGLAKARIPYQKGR 399
Query 60 FPFAANSRARA------TGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYG 113
F+ N R +KVL KILGV ++ +A EL+++A +A+ G
Sbjct 400 VLFSQNVRYNTLLPREENTTVSPFIKVLIDSRDMKILGVHMINDDANELLSQASMAVSLG 459
Query 114 ASSEDLGRVCHAHPTQSEALKEA 136
++ D+ +V HP+ SE+ K+A
Sbjct 460 LTAHDVCKVPFPHPSLSESFKQA 482
> eco:b3962 sthA, ECK3954, JW5551, sth, udhA; pyridine nucleotide
transhydrogenase, soluble (EC:1.6.1.1); K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=466
Score = 68.6 bits (166), Expect = 7e-12, Method: Composition-based stats.
Identities = 49/142 (34%), Positives = 77/142 (54%), Gaps = 5/142 (3%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRH-DTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGA 59
A A ++G + L KG+ T + IP IYT PE++ VGKTE+QL A V Y+ G
Sbjct 319 ASAAYDQGRIAAQALV-KGEATAHLIEDIPTGIYTIPEISSVGKTEQQLTAMKVPYEVGR 377
Query 60 FPFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGEL--IAEAVLAMEYGASS- 116
F +RA+ G + G +K+L +ET +ILG+ G A E+ I +A++ + G ++
Sbjct 378 AQFKHLARAQIVGMNVGTLKILFHRETKEILGIHCFGERAAEIIHIGQAIMEQKGGGNTI 437
Query 117 EDLGRVCHAHPTQSEALKEACM 138
E +PT +EA + A +
Sbjct 438 EYFVNTTFNYPTMAEAYRVAAL 459
> pfa:PF08_0066 lipoamide dehydrogenase, putative (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=666
Score = 65.5 bits (158), Expect = 5e-11, Method: Composition-based stats.
Identities = 44/137 (32%), Positives = 69/137 (50%), Gaps = 23/137 (16%)
Query 23 VRHDTIPAVIYTHPELAGVGKTEEQLKA--------EGVVYKRGA--------------- 59
+ + IP+V YT+PELA +G TE++ K E YK +
Sbjct 520 ILYKNIPSVCYTNPELAFIGLTEKEAKVLYPDNVGVEISYYKSNSKILCENNISLNNNKK 579
Query 60 FPFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDL 119
+ +++G+VK++ ++T +ILG++I+G A LI EAVLA+ S+ DL
Sbjct 580 NNSYNKGQYNINDNTNGMVKIIYKEDTKEILGMFIVGNYASVLIHEAVLAINLKLSAFDL 639
Query 120 GRVCHAHPTQSEALKEA 136
+ H+HPT SE L A
Sbjct 640 AYMVHSHPTVSEVLDTA 656
> eco:b0304 ykgC, ECK0303, JW5040; predicted pyridine nucleotide-disulfide
oxidoreductase
Length=441
Score = 62.4 bits (150), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 37/124 (29%), Positives = 61/124 (49%), Gaps = 1/124 (0%)
Query 13 ERLAGKGD-GTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAFPFAANSRARAT 71
+ L G+G T +P ++ P L+ VG TEEQ + G + P AA RAR
Sbjct 312 DELLGEGKRSTDDRKNVPYSVFMTPPLSRVGMTEEQARESGADIQVVTLPVAAIPRARVM 371
Query 72 GDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQSE 131
D+ G++K + +T ++LG +L ++ E+I + M+ G L HP+ SE
Sbjct 372 NDTRGVLKAIVDNKTQRMLGASLLCVDSHEMINIVKMVMDAGLPYSILRDQIFTHPSMSE 431
Query 132 ALKE 135
+L +
Sbjct 432 SLND 435
> mmu:14782 Gsr, AI325518, D8Ertd238e, Gr-1, Gr1; glutathione
reductase (EC:1.8.1.7); K00383 glutathione reductase (NADPH)
[EC:1.8.1.7]
Length=500
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 33/123 (26%), Positives = 60/123 (48%), Gaps = 12/123 (9%)
Query 18 KGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRG-------AFPFAANSRARA 70
K D + +D IP V+++HP + VG TE++ V+K G + F A
Sbjct 379 KQDSKLDYDNIPTVVFSHPPIGTVGLTEDE-----AVHKYGKDNVKIYSTAFTPMYHAVT 433
Query 71 TGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQS 130
T + ++K++ + +K++G+ + G E++ +A++ GA+ D HPT S
Sbjct 434 TRKTKCVMKMVCANKEEKVVGIHMQGIGCDEMLQGFAVAVKMGATKADFDNTVAIHPTSS 493
Query 131 EAL 133
E L
Sbjct 494 EEL 496
> tgo:TGME49_019130 glutathione reductase, putative (EC:1.8.1.7)
Length=505
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 34/124 (27%), Positives = 57/124 (45%), Gaps = 7/124 (5%)
Query 15 LAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKA----EGVVYKRGAF--PFAANSRA 68
G+ + + +P V+++HP L VG TEE K+ E + F F A
Sbjct 349 FGGRSEARLDLSVVPTVVFSHPALGAVGMTEEDAKSLYGEENINVYTSTFIDSFYAAWSM 408
Query 69 RATGDSDGLVKVLTCKET-DKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHP 127
+ VK++ K DK+LG+ ++G N E++ +A++ GA+ D HP
Sbjct 409 PPSAKPKSFVKMVCLKTANDKVLGLHLVGRNVDEMLQGFAVAIKLGATKADFNSTLAIHP 468
Query 128 TQSE 131
T +E
Sbjct 469 TAAE 472
> dre:100332932 glutathione reductase-like
Length=461
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 54/112 (48%), Gaps = 6/112 (5%)
Query 25 HDTIPAVIYTHPELAGVGKTEE---QLKAEGVVYKRGAFPFAANSRARATGDSDGLVKVL 81
HD I +++ PE+ VG +EE + AE VY+ P A R L+K++
Sbjct 337 HDLIATAVFSQPEIGTVGLSEETAAERYAEIEVYRAEFRPMKATLSGR---QEKMLMKLI 393
Query 82 TCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQSEAL 133
K++G ILG +AGE+ + ++ G + ED R HPT +E L
Sbjct 394 VNAADRKVVGAHILGHDAGEMAQLLGITLKAGCTKEDFDRTMAVHPTAAEEL 445
> xla:734372 gsr, MGC84926; glutathione reductase (EC:1.8.1.7);
K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=476
Score = 51.6 bits (122), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 34/124 (27%), Positives = 62/124 (50%), Gaps = 8/124 (6%)
Query 15 LAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQ-LKAEG----VVYKRGAFPFAANSRAR 69
G+ D + ++ IP V+++HP + VG TEE+ + A+G VY P R
Sbjct 352 FEGQEDSKLDYNNIPTVVFSHPPIGTVGLTEEEAVTAKGRENVKVYTTSFSPMYHVVTRR 411
Query 70 ATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQ 129
T ++K++ + +K++G+ + G E++ +A++ GA+ +D HPT
Sbjct 412 KT---KCVMKLVCVGKEEKVVGLHMQGLGCDEMLQGFAVAIKMGATKKDFDNTVAIHPTS 468
Query 130 SEAL 133
SE L
Sbjct 469 SEEL 472
> cel:ZK637.10 trxr-2; ThioRedoXin Reductase family member (trxr-2);
K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=503
Score = 51.2 bits (121), Expect = 1e-06, Method: Composition-based stats.
Identities = 41/133 (30%), Positives = 57/133 (42%), Gaps = 5/133 (3%)
Query 4 AEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQ-LKAEGV----VYKRG 58
A + G +RL VR D + ++T EL+ VG TEE+ ++ G V+
Sbjct 349 AIQSGKLLADRLFSNSKQIVRFDGVATTVFTPLELSTVGLTEEEAIQKHGEDSIEVFHSH 408
Query 59 AFPFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSED 118
PF + V T E+ KILG+ +GPNA E+I +A G S D
Sbjct 409 FTPFEYVVPQNKDSGFCYVKAVCTRDESQKILGLHFVGPNAAEVIQGYAVAFRVGISMSD 468
Query 119 LGRVCHAHPTQSE 131
L HP SE
Sbjct 469 LQNTIAIHPCSSE 481
> tgo:TGME49_109730 thioredoxin reductase, putative (EC:1.8.1.7);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=662
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 40/138 (28%), Positives = 61/138 (44%), Gaps = 18/138 (13%)
Query 14 RLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEG-------VVYKRGAFPFAANS 66
RL + IP ++T E A G +EE +AE +++ F+
Sbjct 500 RLFANSTEHMDFTNIPTTVFTPIEYAHTGYSEEAAEAEFGRDDLEVYLFQFSPLFFSCVH 559
Query 67 RARATG------DSD----GLVKVLTCK-ETDKILGVWILGPNAGELIAEAVLAMEYGAS 115
R +A D D L K++ K E +K++G+ +GPNAGEL+ LA+ GA
Sbjct 560 REKAPQARKSPEDVDITPPCLAKLICVKSEDEKVVGIHFVGPNAGELMQGFALAVRLGAK 619
Query 116 SEDLGRVCHAHPTQSEAL 133
D + HPT +EA
Sbjct 620 KRDFDKCVGIHPTNAEAF 637
> hsa:10587 TXNRD2, SELZ, TR, TR-BETA, TR3, TRXR2; thioredoxin
reductase 2 (EC:1.8.1.9); K00384 thioredoxin reductase (NADPH)
[EC:1.8.1.9]
Length=524
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 43/130 (33%), Positives = 61/130 (46%), Gaps = 8/130 (6%)
Query 8 GIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKA----EGV-VYKRGAFPF 62
G V+RL G + +D +P ++T E VG +EE+ A E V VY P
Sbjct 375 GRLLVQRLFGGSSDLMDYDNVPTTVFTPLEYGCVGLSEEEAVARHGQEHVEVYHAHYKPL 434
Query 63 AANSRARATGDSDGLVKVLTCKETDK-ILGVWILGPNAGELIAEAVLAMEYGASSEDLGR 121
R S VK++ +E + +LG+ LGPNAGE+ L ++ GAS + R
Sbjct 435 EFTVAGRDA--SQCYVKMVCLREPPQLVLGLHFLGPNAGEVTQGFALGIKCGASYAQVMR 492
Query 122 VCHAHPTQSE 131
HPT SE
Sbjct 493 TVGIHPTCSE 502
> hsa:2936 GSR, MGC78522; glutathione reductase (EC:1.8.1.7);
K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=522
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/118 (26%), Positives = 58/118 (49%), Gaps = 2/118 (1%)
Query 18 KGDGTVRHDTIPAVIYTHPELAGVGKTE-EQLKAEGV-VYKRGAFPFAANSRARATGDSD 75
K D + ++ IP V+++HP + VG TE E + G+ K + F A +
Sbjct 401 KEDSKLDYNNIPTVVFSHPPIGTVGLTEDEAIHKYGIENVKTYSTSFTPMYHAVTKRKTK 460
Query 76 GLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQSEAL 133
++K++ + +K++G+ + G E++ +A++ GA+ D HPT SE L
Sbjct 461 CVMKMVCANKEEKVVGIHMQGLGCDEMLQGFAVAVKMGATKADFDNTVAIHPTSSEEL 518
> sce:YPL091W GLR1, LPG17; Cytosolic and mitochondrial glutathione
oxidoreductase, converts oxidized glutathione to reduced
glutathione; mitochondrial but not cytosolic form has a role
in resistance to hyperoxia (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=483
Score = 49.7 bits (117), Expect = 4e-06, Method: Composition-based stats.
Identities = 31/114 (27%), Positives = 58/114 (50%), Gaps = 8/114 (7%)
Query 25 HDTIPAVIYTHPELAGVGKTE----EQLKAEGV-VYKRGAFPFAANSRARATGDSDGLVK 79
++ +P+VI++HPE +G +E E+ E + VY F A A + S K
Sbjct 369 YENVPSVIFSHPEAGSIGISEKEAIEKYGKENIKVYNS---KFTAMYYAMLSEKSPTRYK 425
Query 80 VLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQSEAL 133
++ +K++G+ I+G ++ E++ +A++ GA+ D HPT +E L
Sbjct 426 IVCAGPNEKVVGLHIVGDSSAEILQGFGVAIKMGATKADFDNCVAIHPTSAEEL 479
> eco:b3500 gor, ECK3485, gorA, JW3467; glutathione oxidoreductase
(EC:1.8.1.7); K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=450
Score = 48.5 bits (114), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 36/127 (28%), Positives = 59/127 (46%), Gaps = 9/127 (7%)
Query 13 ERL-AGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEG-----VVYKRGAFPFAANS 66
ERL K D + + IP V+++HP + VG TE Q + + VYK F A
Sbjct 323 ERLFNNKPDEHLDYSNIPTVVFSHPPIGTVGLTEPQAREQYGDDQVKVYKSS---FTAMY 379
Query 67 RARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAH 126
A T +K++ +KI+G+ +G E++ +A++ GA+ +D H
Sbjct 380 TAVTTHRQPCRMKLVCVGSEEKIVGIHGIGFGMDEMLQGFAVALKMGATKKDFDNTVAIH 439
Query 127 PTQSEAL 133
PT +E
Sbjct 440 PTAAEEF 446
> cel:C06G3.7 trxr-1; ThioRedoXin Reductase family member (trxr-1);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=667
Score = 48.1 bits (113), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 34/131 (25%), Positives = 53/131 (40%), Gaps = 5/131 (3%)
Query 8 GIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQL-----KAEGVVYKRGAFPF 62
G + R+ + +D IP ++T E G +EE K ++Y P
Sbjct 516 GRVLMRRIFDGANELTEYDQIPTTVFTPLEYGCCGLSEEDAMMKYGKDNIIIYHNVFNPL 575
Query 63 AANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRV 122
R D L + E +K++G IL PNAGE+ +A++ A D R+
Sbjct 576 EYTISERMDKDHCYLKMICLRNEEEKVVGFHILTPNAGEVTQGFGIALKLAAKKADFDRL 635
Query 123 CHAHPTQSEAL 133
HPT +E
Sbjct 636 IGIHPTVAENF 646
> bbo:BBOV_I002190 19.m02214; thiodoxin reductase (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=559
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 39/144 (27%), Positives = 66/144 (45%), Gaps = 14/144 (9%)
Query 4 AEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQ-LKAEGVV--YKR--G 58
A ++G RL G + + + +P ++T E G +EE+ +K G V Y +
Sbjct 391 AVKDGEMLARRLFGNSNKLMDLNYVPMCVFTPIEYGKCGLSEEEAVKKYGDVDIYLKEFT 450
Query 59 AFPFAANSRARA----TGDSD-----GLVKVLTCKETDKILGVWILGPNAGELIAEAVLA 109
+ F+A R + T + D + + CK+ I+G+ +GPNAGE+I +A
Sbjct 451 SLEFSAVHRHKVEWMQTDEMDVDMPPTCLSKMICKKDGTIVGIHFVGPNAGEIIQGLCVA 510
Query 110 MEYGASSEDLGRVCHAHPTQSEAL 133
+ GA D HPT +E+
Sbjct 511 VRLGAKKSDFDDTIGVHPTDAESF 534
> tgo:TGME49_046920 glutathione reductase, putative (EC:1.8.1.7);
K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=483
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 55/113 (48%), Gaps = 15/113 (13%)
Query 28 IPAVIYTHPELAGVGKTEEQLKAE--------GVVYKRGAFPFAANSRARATGDSDGLVK 79
+P V+++HP A VG TE + KA V GA+P A + + +K
Sbjct 362 VPTVVFSHPPAACVGLTEAEAKATYGEKDIKVHVNLYYGAWPVAPEEKPKT------FIK 415
Query 80 VLTCK-ETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQSE 131
++ K + K++G+ ++G A E+I +AM+ GA+ D HPT +E
Sbjct 416 MICVKSQMLKVVGLHVVGMGADEMIQGFGVAMKMGATKADFDNCVAVHPTAAE 468
> ath:AT3G54660 GR; GR (GLUTATHIONE REDUCTASE); ATP binding /
glutathione-disulfide reductase (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=565
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 31/111 (27%), Positives = 54/111 (48%), Gaps = 8/111 (7%)
Query 25 HDTIPAVIYTHPELAGVGKTEEQL---KAEGVVYKRGAFPFAANSRARATGDSDG-LVKV 80
+ +P +++ P + VG TEEQ + VY P +A +G D +K+
Sbjct 428 YRAVPCAVFSQPPIGTVGLTEEQAIEQYGDVDVYTSNFRPL----KATLSGLPDRVFMKL 483
Query 81 LTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQSE 131
+ C T+K+LGV + G ++ E+I +A++ G + D HPT +E
Sbjct 484 IVCANTNKVLGVHMCGEDSPEIIQGFGVAVKAGLTKADFDATVGVHPTAAE 534
> xla:447610 txnrd1, MGC85342; thioredoxin reductase 1 (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=531
Score = 45.8 bits (107), Expect = 5e-05, Method: Composition-based stats.
Identities = 34/122 (27%), Positives = 57/122 (46%), Gaps = 12/122 (9%)
Query 19 GDGTVRHD--TIPAVIYTHPELAGVGKTEE----QLKAEGV-VYKRGAFPFAANSRARAT 71
GD T++ D +P ++T E G +EE Q E V VY +P AR
Sbjct 393 GDSTLKSDYVNVPTTVFTPLEYGACGLSEENAIRQYGEENVEVYHSYFWPLEWTVPAR-- 450
Query 72 GDSDGLVKVLTC--KETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQ 129
D++ + C K+ ++++G +L PNAGE+ +A++ G + + L HP
Sbjct 451 -DNNKCYAKIICNLKDNERVVGFHVLSPNAGEITQGFAVAIKCGLTKDQLDNTIGIHPVC 509
Query 130 SE 131
+E
Sbjct 510 AE 511
> mmu:26462 Txnrd2, AA118373, ESTM573010, TGR, Tr3, Trxr2, Trxrd2;
thioredoxin reductase 2 (EC:1.8.1.9); K00384 thioredoxin
reductase (NADPH) [EC:1.8.1.9]
Length=527
Score = 45.4 bits (106), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 39/125 (31%), Positives = 59/125 (47%), Gaps = 8/125 (6%)
Query 13 ERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKA----EGV-VYKRGAFPFAANSR 67
+RL GK + + +P ++T E VG +EE+ A E V VY P
Sbjct 383 QRLFGKSSTLMDYSNVPTTVFTPLEYGCVGLSEEEAVALHGQEHVEVYHAYYKPLEFTVA 442
Query 68 ARATGDSDGLVKVLTCKETDK-ILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAH 126
R S +K++ +E + +LG+ LGPNAGE+ L ++ GAS + + H
Sbjct 443 DRDA--SQCYIKMVCMREPPQLVLGLHFLGPNAGEVTQGFALGIKCGASYAQVMQTVGIH 500
Query 127 PTQSE 131
PT SE
Sbjct 501 PTCSE 505
> cpv:cgd2_4320 thioredoxin reductase 1 ; K00384 thioredoxin reductase
(NADPH) [EC:1.8.1.9]
Length=526
Score = 45.1 bits (105), Expect = 8e-05, Method: Composition-based stats.
Identities = 37/148 (25%), Positives = 63/148 (42%), Gaps = 18/148 (12%)
Query 4 AEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKA-------EGVVYK 56
A + GI RL + + +D +P ++T E VG + E A E + +
Sbjct 354 AVKAGILLARRLFAGSNEFIDYDFVPTTVFTPIEYGHVGLSSEAAIAKYGEDDIEEYLSE 413
Query 57 RGAFPFAANSRARATGDSDG----------LVKVLTCK-ETDKILGVWILGPNAGELIAE 105
AA R + + L K++ K + +K++G +GPNAGE+
Sbjct 414 FSTLEIAAAHREKPEHLRENEMDFALPLNCLAKLVVVKSQGEKVVGFHFVGPNAGEITQG 473
Query 106 AVLAMEYGASSEDLGRVCHAHPTQSEAL 133
LA++ GA+ +D + HPT +E
Sbjct 474 FSLAVKLGATKKDFDDMIGIHPTDAEVF 501
> mmu:50493 Txnrd1, TR, TR1, TrxR1; thioredoxin reductase 1 (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=499
Score = 45.1 bits (105), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 34/137 (24%), Positives = 61/137 (44%), Gaps = 10/137 (7%)
Query 4 AEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQ----LKAEGV-VYKRG 58
A + G +RL G + +D +P ++T E G +EE+ E + VY
Sbjct 346 AIQAGRLLAQRLYGGSNVKCDYDNVPTTVFTPLEYGCCGLSEEKAVEKFGEENIEVYHSF 405
Query 59 AFPFAANSRARATGDSDGLVKVLTC--KETDKILGVWILGPNAGELIAEAVLAMEYGASS 116
+P +R D++ + C K+ ++++G +LGPNAGE+ A++ G +
Sbjct 406 FWPLEWTVPSR---DNNKCYAKIICNLKDDERVVGFHVLGPNAGEVTQGFAAALKCGLTK 462
Query 117 EDLGRVCHAHPTQSEAL 133
+ L HP +E
Sbjct 463 QQLDSTIGIHPVCAEIF 479
> ath:AT3G24170 ATGR1; ATGR1 (glutathione-disulfide reductase);
FAD binding / NADP or NADPH binding / glutathione-disulfide
reductase/ oxidoreductase (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=499
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/111 (24%), Positives = 53/111 (47%), Gaps = 7/111 (6%)
Query 25 HDTIPAVIYTHPELAGVGKTEE----QLKAEGVVYKRGAFPFAANSRARATGDSDGLVKV 80
+ + ++ P LA VG +EE Q + +V+ G P R L+K+
Sbjct 370 YSNVACAVFCIPPLAVVGLSEEEAVEQATGDILVFTSGFNPMKNTISGR---QEKTLMKL 426
Query 81 LTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQSE 131
+ +++DK++G + GP+A E++ +A++ GA+ HP+ +E
Sbjct 427 IVDEKSDKVIGASMCGPDAAEIMQGIAIALKCGATKAQFDSTVGIHPSSAE 477
> xla:379744 txnrd2, MGC69182; thioredoxin reductase 2 (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=504
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 33/117 (28%), Positives = 55/117 (47%), Gaps = 16/117 (13%)
Query 25 HDTIPAVIYTHPELAGVGKTEEQLKAE---------GVVYKRGAFPFAANSRARATGDSD 75
+D++P ++T E VG +EE+ K YK F A ++
Sbjct 372 YDSVPTTVFTPLEYGCVGISEEEAKERYGDDNIEVFHAFYKPLEFTVAERDASQC----- 426
Query 76 GLVKVLTCKETD-KILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQSE 131
+K++ ++ D +ILG+ + GPNAGE+I L ++ GA+ L HPT +E
Sbjct 427 -YIKIICLRKHDQRILGLHLTGPNAGEVIQGFALGIKCGATYPQLMCTVGIHPTCAE 482
> tpv:TP03_0110 thioredoxin reductase (EC:1.6.4.5); K00384 thioredoxin
reductase (NADPH) [EC:1.8.1.9]
Length=567
Score = 42.7 bits (99), Expect = 4e-04, Method: Composition-based stats.
Identities = 34/135 (25%), Positives = 58/135 (42%), Gaps = 14/135 (10%)
Query 12 VERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQL-------KAEGVVYKRGAFPFAA 64
++RL + + ++ +P +YT E + G TEE+ E + + +
Sbjct 409 IQRLYSNTNTKMNYENVPKCVYTPFEYSSCGLTEEEAIERFGEENLEIYLKEYNNLEISP 468
Query 65 NSRA-RATGDS-----DGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSED 118
R + T D L KV+ C KI+G+ +GPNAGE++ + A D
Sbjct 469 VHRINKKTNDEFDYPMTCLSKVI-CLRDGKIVGMHFVGPNAGEIMQGFSVLFTLNAKKSD 527
Query 119 LGRVCHAHPTQSEAL 133
L + HPT +E+
Sbjct 528 LDKTVGIHPTDAESF 542
> pfa:PFI1170c thioredoxin reductase (EC:1.8.1.9); K00384 thioredoxin
reductase (NADPH) [EC:1.8.1.9]
Length=617
Score = 42.4 bits (98), Expect = 6e-04, Method: Composition-based stats.
Identities = 38/148 (25%), Positives = 56/148 (37%), Gaps = 18/148 (12%)
Query 4 AEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQL-------KAEGVVYK 56
A + G RL D + + IP IYT E G +EE+ E + +
Sbjct 445 AIKAGEILARRLFKDSDEIMDYSYIPTSIYTPIEYGACGYSEEKAYELYGKSNVEVFLQE 504
Query 57 RGAFPFAANSRA---RATGDSDGLVKVLTC--------KETDKILGVWILGPNAGELIAE 105
+A R RA D L TC E ++++G +GPNAGE+
Sbjct 505 FNNLEISAVHRQKHIRAQKDEYDLDVSSTCLAKLVCLKNEDNRVIGFHYVGPNAGEVTQG 564
Query 106 AVLAMEYGASSEDLGRVCHAHPTQSEAL 133
LA+ +D HPT +E+
Sbjct 565 MALALRLKVKKKDFDNCIGIHPTDAESF 592
> mmu:232223 Txnrd3, AI196535, TR2, Tgr; thioredoxin reductase
3 (EC:1.8.1.9); K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=652
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 37/137 (27%), Positives = 57/137 (41%), Gaps = 10/137 (7%)
Query 4 AEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQ----LKAEGV-VYKRG 58
A + G RL G + IP ++T E G +EE+ K E + VY
Sbjct 499 AIQAGKLLARRLFGVSLEKCDYINIPTTVFTPLEYGCCGLSEEKAIEMYKKENLEVYHTL 558
Query 59 AFPFAANSRARATGDSDGLVKVLTCKETD--KILGVWILGPNAGELIAEAVLAMEYGASS 116
+P R D++ + C + D +++G +LGPNAGE+ AM+ G +
Sbjct 559 FWPLEWTVAGR---DNNTCYAKIICNKFDNERVVGFHLLGPNAGEITQGFAAAMKCGLTK 615
Query 117 EDLGRVCHAHPTQSEAL 133
+ L HPT E
Sbjct 616 QLLDDTIGIHPTCGEVF 632
> xla:447484 txnrd3, MGC81848; thioredoxin reductase 3 (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=596
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 31/114 (27%), Positives = 51/114 (44%), Gaps = 10/114 (8%)
Query 27 TIPAVIYTHPELAGVGKTEEQ----LKAEGV-VYKRGAFPFAANSRARATGDSDGLVKVL 81
+P ++T E G EE+ E + VY +P +R D++ +
Sbjct 468 NVPTTVFTPLEYGCCGYAEEKAIEIYGEENLEVYHTLFWPLEWTVPSR---DNNTCFAKI 524
Query 82 TC--KETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQSEAL 133
C ++ D+++G +LGPNAGE+ AM+ G + E L HPT +E
Sbjct 525 ICNKQDNDRVIGFHVLGPNAGEITQGFGAAMKCGLTKEKLDETIGIHPTCAEIF 578
> pfa:PF14_0192 glutathione reductase (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=500
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 31/125 (24%), Positives = 54/125 (43%), Gaps = 6/125 (4%)
Query 13 ERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQ----LKAEGVVYKRGAFP--FAANS 66
+RL K + IP VI++HP + +G +EE E V F F +
Sbjct 366 DRLFLKKTRKTNYKLIPTVIFSHPPIGTIGLSEEAAIQIYGKENVKIYESKFTNLFFSVY 425
Query 67 RARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAH 126
+K++ + + I G+ I+G NA E++ +A++ A+ +D H
Sbjct 426 DIEPELKEKTYLKLVCVGKDELIKGLHIIGLNADEIVQGFAVALKMNATKKDFDETIPIH 485
Query 127 PTQSE 131
PT +E
Sbjct 486 PTAAE 490
> hsa:7296 TXNRD1, GRIM-12, MGC9145, TR, TR1, TRXR1, TXNR; thioredoxin
reductase 1 (EC:1.8.1.9); K00384 thioredoxin reductase
(NADPH) [EC:1.8.1.9]
Length=649
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 32/137 (23%), Positives = 59/137 (43%), Gaps = 10/137 (7%)
Query 4 AEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQ----LKAEGV-VYKRG 58
A + G +RL ++ +P ++T E G +EE+ E + VY
Sbjct 496 AIQAGRLLAQRLYAGSTVKCDYENVPTTVFTPLEYGACGLSEEKAVEKFGEENIEVYHSY 555
Query 59 AFPFAANSRARATGDSDGLVKVLTC--KETDKILGVWILGPNAGELIAEAVLAMEYGASS 116
+P +R D++ + C K+ ++++G +LGPNAGE+ A++ G +
Sbjct 556 FWPLEWTIPSR---DNNKCYAKIICNTKDNERVVGFHVLGPNAGEVTQGFAAALKCGLTK 612
Query 117 EDLGRVCHAHPTQSEAL 133
+ L HP +E
Sbjct 613 KQLDSTIGIHPVCAEVF 629
> dre:352924 txnrd1, TrxR1, cb682, fb83a08, wu:fb83a08; thioredoxin
reductase 1 (EC:1.8.1.9); K00384 thioredoxin reductase
(NADPH) [EC:1.8.1.9]
Length=602
Score = 38.5 bits (88), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 49/114 (42%), Gaps = 10/114 (8%)
Query 27 TIPAVIYTHPELAGVGKTEEQ-LKAEGV----VYKRGAFPFAANSRARATGDSDGLVKVL 81
+P ++T E G EE+ ++ G VY +P R D++ +
Sbjct 472 NVPTTVFTPMEYGSCGHPEEKAIQMYGQENLEVYHSLFWPLEFTVPGR---DNNKCYAKI 528
Query 82 TCKETD--KILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQSEAL 133
C + D +++G LGPNAGE+ AM+ G + + L HPT +E
Sbjct 529 ICNKLDNLRVIGFHYLGPNAGEVTQGFGAAMKCGITKDQLDNTIGIHPTCAEIF 582
> dre:553575 MGC110010; zgc:110010 (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=425
Score = 36.2 bits (82), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 39/82 (47%), Gaps = 7/82 (8%)
Query 54 VYKRGAFP--FAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAME 111
VY P +A SR S ++K++ E +K++G+ + G E++ +A+
Sbjct 345 VYTTSFTPMYYAITSRK-----SQCIMKLVCAGENEKVVGLHMQGFGCDEMLQGFAVAVN 399
Query 112 YGASSEDLGRVCHAHPTQSEAL 133
GA+ D R HPT SE L
Sbjct 400 MGATKADFDRTIAIHPTSSEEL 421
> hsa:114112 TXNRD3, TGR, TR2, TRXR3; thioredoxin reductase 3
(EC:1.8.1.9); K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=607
Score = 35.8 bits (81), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 87 DKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQSEAL 133
D+++G ILGPNAGE+ AM+ G + + L HPT E
Sbjct 541 DRVIGFHILGPNAGEVTQGFAAAMKCGLTKQLLDDTIGIHPTCGEVF 587
> cel:C46F11.2 hypothetical protein
Length=473
Score = 33.1 bits (74), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 31/126 (24%), Positives = 58/126 (46%), Gaps = 12/126 (9%)
Query 15 LAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQL-----KAEGVVYKRGAFP--FAANSR 67
G+ D + ++ I V+++HP + VG TE + K E +YK P FA
Sbjct 343 FNGETDNKLTYENIATVVFSHPLIGTVGLTEAEAVEKYGKDEVTLYKSRFNPMLFAVTKH 402
Query 68 ARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHP 127
+K++ + +K++GV + G + E++ +A+ GA+ + + HP
Sbjct 403 KEKAA-----MKLVCVGKDEKVVGVHVFGVGSDEMLQGFAVAVTMGATKKQFDQTVAIHP 457
Query 128 TQSEAL 133
T +E L
Sbjct 458 TSAEEL 463
Lambda K H
0.316 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3003468616
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40