bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4425_orf1
Length=112
Score E
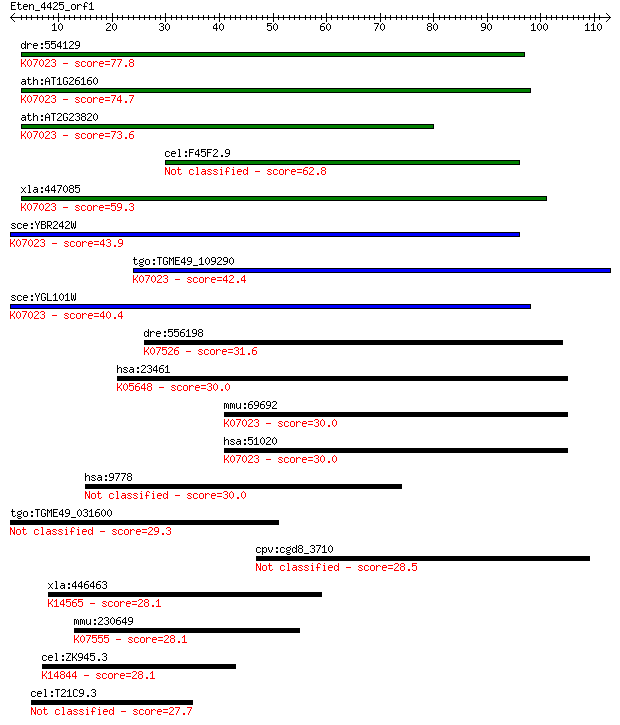
Sequences producing significant alignments: (Bits) Value
dre:554129 hddc2, fb71h08, wu:fb71h08, zgc:112330; HD domain c... 77.8 7e-15
ath:AT1G26160 metal-dependent phosphohydrolase HD domain-conta... 74.7 6e-14
ath:AT2G23820 metal-dependent phosphohydrolase HD domain-conta... 73.6 2e-13
cel:F45F2.9 hypothetical protein 62.8 3e-10
xla:447085 hddc2, MGC85244; HD domain containing 2; K07023 put... 59.3 3e-09
sce:YBR242W Putative protein of unknown function; green fluore... 43.9 1e-04
tgo:TGME49_109290 HD domain-containing protein ; K07023 putati... 42.4 3e-04
sce:YGL101W Putative protein of unknown function; non-essentia... 40.4 0.001
dre:556198 srgap2a, MGC136814, srgap2, wu:fb47h12, zgc:136814;... 31.6 0.73
hsa:23461 ABCA5, ABC13, DKFZp451F117, DKFZp779N2435, EST90625,... 30.0 1.7
mmu:69692 Hddc2, 2310057G13Rik, MGC129436, MGC129437; HD domai... 30.0 1.7
hsa:51020 HDDC2, C6orf74, MGC87330, NS5ATP2, dJ167O5.2; HD dom... 30.0 1.8
hsa:9778 KIAA0232 30.0 1.9
tgo:TGME49_031600 importin, putative 29.3 3.1
cpv:cgd8_3710 hypothetical protein 28.5 6.1
xla:446463 nop58-a, MGC78950, nop5, nop5/nop58, nop58; NOP58 r... 28.1 6.3
mmu:230649 Atpaf1, 6330547J17Rik, AI593252, ATP11, ATP11p; ATP... 28.1 6.8
cel:ZK945.3 puf-12; PUF (Pumilio/FBF) domain-containing family... 28.1 7.4
cel:T21C9.3 hypothetical protein 27.7 9.6
> dre:554129 hddc2, fb71h08, wu:fb71h08, zgc:112330; HD domain
containing 2; K07023 putative hydrolases of HD superfamily
Length=200
Score = 77.8 bits (190), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 43/96 (44%), Positives = 56/96 (58%), Gaps = 2/96 (2%)
Query 3 EKREKEKNAFLFICKFLDDKRAKEVRELWEEYERGETPEAMLVKDADKFDMITKAFEYEN 62
EK +EK+A + I LDD KE+ LWEEYE +PEA LVK+ D +MI +A EYE
Sbjct 84 EKHRREKDAMVHITGLLDDGLRKEIYNLWEEYETQSSPEAKLVKELDNLEMIIQAHEYEE 143
Query 63 SHGVC--LEEFFSSTENAFRTEVFKELNRSLRQQRS 96
G L+EFF STE F L +SL ++R+
Sbjct 144 LEGKPGRLQEFFVSTEGKFHHPEVLGLLKSLNEERA 179
> ath:AT1G26160 metal-dependent phosphohydrolase HD domain-containing
protein; K07023 putative hydrolases of HD superfamily
Length=258
Score = 74.7 bits (182), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 42/96 (43%), Positives = 56/96 (58%), Gaps = 1/96 (1%)
Query 3 EKREKEKNAFLFICKFLDDK-RAKEVRELWEEYERGETPEAMLVKDADKFDMITKAFEYE 61
EK +E A +C+ L RA+E+ ELW EYE + EA +VKD DK +MI +A EYE
Sbjct 156 EKSRRETAALKEMCEVLGGGLRAEEITELWLEYENNASLEANIVKDFDKVEMILQALEYE 215
Query 62 NSHGVCLEEFFSSTENAFRTEVFKELNRSLRQQRSS 97
HG L+EFF ST F+TE+ K + +R S
Sbjct 216 AEHGKVLDEFFISTAGKFQTEIGKSWAAEINARRKS 251
> ath:AT2G23820 metal-dependent phosphohydrolase HD domain-containing
protein; K07023 putative hydrolases of HD superfamily
Length=245
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 39/78 (50%), Positives = 48/78 (61%), Gaps = 1/78 (1%)
Query 3 EKREKEKNAFLFICKFLDD-KRAKEVRELWEEYERGETPEAMLVKDADKFDMITKAFEYE 61
EK +E A +CK L +RAKE+ ELW EYE +PEA +VKD DK ++I +A EYE
Sbjct 160 EKNRRESEALEHMCKLLGGGERAKEIAELWREYEENSSPEAKVVKDFDKVELILQALEYE 219
Query 62 NSHGVCLEEFFSSTENAF 79
G LEEFF ST F
Sbjct 220 QDQGKDLEEFFQSTAGNF 237
> cel:F45F2.9 hypothetical protein
Length=75
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 42/66 (63%), Gaps = 0/66 (0%)
Query 30 LWEEYERGETPEAMLVKDADKFDMITKAFEYENSHGVCLEEFFSSTENAFRTEVFKELNR 89
LW+EYE + A +VK DKFDMI +A +YE +H + L++FF+ST + E F +R
Sbjct 2 LWKEYEEASSLTARVVKHLDKFDMIVQADKYEKTHEIDLQQFFTSTVGVLKMEPFATWDR 61
Query 90 SLRQQR 95
LR+ R
Sbjct 62 ELRENR 67
> xla:447085 hddc2, MGC85244; HD domain containing 2; K07023 putative
hydrolases of HD superfamily
Length=201
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 36/100 (36%), Positives = 55/100 (55%), Gaps = 2/100 (2%)
Query 3 EKREKEKNAFLFICKFLDDKRAKEVRELWEEYERGETPEAMLVKDADKFDMITKAFEYE- 61
EK KEK A + + L D EV +LWEEYE T EA VK+ D+ +MI +A EYE
Sbjct 94 EKHRKEKAAMEHLTQLLPDNLKTEVYDLWEEYEHQFTAEAKFVKELDQCEMILQALEYEE 153
Query 62 -NSHGVCLEEFFSSTENAFRTEVFKELNRSLRQQRSSCCS 100
L++F++ST F+ +L ++ ++R+S +
Sbjct 154 LEKRPGRLQDFYNSTAGKFKHPEIVQLVSAIYEERNSAIA 193
> sce:YBR242W Putative protein of unknown function; green fluorescent
protein (GFP)-fusion protein localizes to the cytoplasm
and nucleus; YBR242W is not an essential gene; K07023 putative
hydrolases of HD superfamily
Length=238
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 32/99 (32%), Positives = 51/99 (51%), Gaps = 5/99 (5%)
Query 1 EKEKREKEKNAFL--FICKFLDDKRAKEVRELWEEYERGETPEAMLVKDADKFDMITKAF 58
EK +RE E +L + K ++ AKE+ + W YE + EA VKD DK++M+ + F
Sbjct 128 EKHRREWETIKYLCNALIKPYNEIAAKEIMDDWLAYENVTSLEARYVKDIDKYEMLVQCF 187
Query 59 EYENSHGVC--LEEFFSSTENAFRTEVFKELNRSLRQQR 95
EYE + ++FF + + +T+ K L QR
Sbjct 188 EYEREYKGTKNFDDFFGAVA-SIKTDEVKGWTSDLVVQR 225
> tgo:TGME49_109290 HD domain-containing protein ; K07023 putative
hydrolases of HD superfamily
Length=305
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 40/129 (31%), Positives = 56/129 (43%), Gaps = 40/129 (31%)
Query 24 AKEVRELWEEYERGETPEAMLVKDADKFDMITKAFEYEN-------SHG----------- 65
+E+ LWEEYE G T EA V D DKF+MI +AFEYE+ S G
Sbjct 172 GEEILSLWEEYEEGTTEEAKYVFDIDKFEMILQAFEYESDPSQEGPSSGPFYHRAQEETE 231
Query 66 ----------------------VCLEEFFSSTENAFRTEVFKELNRSLRQQRSSCCSISS 103
+ + F+ STEN FR+++FK L+ LR +R + +
Sbjct 232 EKEHQHHTEAAAEENGSRKRRRLYMPSFYRSTENVFRSDLFKSLDFVLRNRRGTLPQVQR 291
Query 104 SCCCCVQKQ 112
+ Q Q
Sbjct 292 AAKLEAQNQ 300
> sce:YGL101W Putative protein of unknown function; non-essential
gene with similarity to YBR242W; interacts with the DNA
helicase Hpr5p; K07023 putative hydrolases of HD superfamily
Length=215
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 30/101 (29%), Positives = 53/101 (52%), Gaps = 5/101 (4%)
Query 1 EKEKREKEKNAFL--FICKFLDDKRAKEVRELWEEYERGETPEAMLVKDADKFDMITKAF 58
EK +RE E +L I + + ++E+ + W YE+ E VKD DK++M+ + F
Sbjct 109 EKHRREFETVKYLCESIIRPCSESASREILDDWLAYEKQTCLEGRYVKDIDKYEMLVQCF 168
Query 59 EYENSHGV--CLEEFFSSTENAFRTEVFKELNRSLRQQRSS 97
EYE + L++F + N +T+ K+ +SL + R +
Sbjct 169 EYEQKYNGKKDLKQFLGAI-NDIKTDEVKKWTQSLLEDRQA 208
> dre:556198 srgap2a, MGC136814, srgap2, wu:fb47h12, zgc:136814;
SLIT-ROBO Rho GTPase activating protein 2a; K07526 SLIT-ROBO
Rho GTPase activating protein
Length=1100
Score = 31.6 bits (70), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 37/79 (46%), Gaps = 20/79 (25%)
Query 26 EVRELWEEYERGETPEAMLVKDADKFDMITKAFEYENSHGVCLEEFFSSTENA-FRTEVF 84
EV ++ +ERGE P L D + DM + A GV L+ +F ENA F EVF
Sbjct 535 EVNDIKNAFERGEDP---LAGDQNDHDMDSIA-------GV-LKLYFRGLENALFPKEVF 583
Query 85 KELNRSLRQQRSSCCSISS 103
+L SC SI S
Sbjct 584 HDL--------MSCVSIES 594
> hsa:23461 ABCA5, ABC13, DKFZp451F117, DKFZp779N2435, EST90625,
FLJ16381; ATP-binding cassette, sub-family A (ABC1), member
5; K05648 ATP-binding cassette, subfamily A (ABC1), member
5
Length=1642
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 20/88 (22%), Positives = 44/88 (50%), Gaps = 6/88 (6%)
Query 21 DKRAKEVRELWEEYERGETPEAMLVKDADKFDMITKAFEY----ENSHGVCLEEFFSSTE 76
D+ +E++ ++ R E+ ++L K D+ + + + E H +EE+ S
Sbjct 1544 DRLQREIQYIFPNASRQESFSSILAYKIPKEDVQSLSQSFFKLEEAKHAFAIEEY--SFS 1601
Query 77 NAFRTEVFKELNRSLRQQRSSCCSISSS 104
A +VF EL + ++ +SC +++S+
Sbjct 1602 QATLEQVFVELTKEQEEEDNSCGTLNST 1629
> mmu:69692 Hddc2, 2310057G13Rik, MGC129436, MGC129437; HD domain
containing 2; K07023 putative hydrolases of HD superfamily
Length=199
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 35/67 (52%), Gaps = 4/67 (5%)
Query 41 EAMLVKDADKFDMITKAFEY---ENSHGVCLEEFFSSTENAFRTEVFKELNRSLRQQRSS 97
EA VK D+ +MI +A EY EN G L++F+ ST F +L L +R++
Sbjct 130 EAKFVKQLDQCEMILQASEYEDLENKPG-RLQDFYDSTAGKFSHPEIVQLVSELETERNA 188
Query 98 CCSISSS 104
+ +S+
Sbjct 189 SMATASA 195
> hsa:51020 HDDC2, C6orf74, MGC87330, NS5ATP2, dJ167O5.2; HD domain
containing 2; K07023 putative hydrolases of HD superfamily
Length=204
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 35/66 (53%), Gaps = 2/66 (3%)
Query 41 EAMLVKDADKFDMITKAFEYEN-SHGVC-LEEFFSSTENAFRTEVFKELNRSLRQQRSSC 98
EA VK D+ +MI +A EYE+ H L++F+ ST F +L L +RS+
Sbjct 135 EAKFVKQLDQCEMILQASEYEDLEHKPGRLQDFYDSTAGKFNHPEIVQLVSELEAERSTN 194
Query 99 CSISSS 104
+ ++S
Sbjct 195 IAAAAS 200
> hsa:9778 KIAA0232
Length=1395
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 33/60 (55%), Gaps = 1/60 (1%)
Query 15 ICKFLDDKRAKEVRELWEEYERGETPEAMLVKDA-DKFDMITKAFEYENSHGVCLEEFFS 73
+C L D ++K+ ++ ++ E +PEA L A D+ +M +AF + VCL+E +
Sbjct 134 LCSRLKDLQSKQEEKIHKKLEGSPSPEAELSPPAKDQVEMYYEAFPPLSEKPVCLQEIMT 193
> tgo:TGME49_031600 importin, putative
Length=1169
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 30/50 (60%), Gaps = 0/50 (0%)
Query 1 EKEKREKEKNAFLFICKFLDDKRAKEVRELWEEYERGETPEAMLVKDADK 50
++E+ E+ + A+L + K + D R +EVR+L R + +AM V D ++
Sbjct 43 QQEQVEQRRLAYLLLAKQIVDGRQEEVRQLSAVLLRRKISKAMAVFDENQ 92
> cpv:cgd8_3710 hypothetical protein
Length=401
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 17/68 (25%), Positives = 26/68 (38%), Gaps = 9/68 (13%)
Query 47 DADKFDMITKAFEYENSHGVCLEEFFSSTENAFRTEVFKELNRSLRQ------QRSSCCS 100
D +++K EN HG E NA R E + R+++ + CC+
Sbjct 185 DGSARSLLSKYINGENKHG---NEVREKDPNAIRREKVDNIKRTVKPIARDNGNKGKCCA 241
Query 101 ISSSCCCC 108
CCC
Sbjct 242 FWKKWCCC 249
> xla:446463 nop58-a, MGC78950, nop5, nop5/nop58, nop58; NOP58
ribonucleoprotein homolog; K14565 nucleolar protein 58
Length=534
Score = 28.1 bits (61), Expect = 6.3, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 28/52 (53%), Gaps = 1/52 (1%)
Query 8 EKNAFLFICKFLDDKRAKEVRELWEEYERGETPEAML-VKDADKFDMITKAF 58
E A I K LD+ + +EV +W+E+E E ++ +K +KF T+A
Sbjct 6 ETAAGYAIFKVLDESKLQEVDSIWKEFETPEKANKVVKLKHFEKFQDTTEAL 57
> mmu:230649 Atpaf1, 6330547J17Rik, AI593252, ATP11, ATP11p; ATP
synthase mitochondrial F1 complex assembly factor 1; K07555
ATP synthase mitochondrial F1 complex assembly factor 1
Length=324
Score = 28.1 bits (61), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 29/43 (67%), Gaps = 3/43 (6%)
Query 13 LFICKFLDDKRAKEVRELWEEY-ERGETPEAMLVKDADKFDMI 54
+F + + DK A+E++++W++Y +T A++ K +KFD+I
Sbjct 143 VFNVEMVKDKTAEEIKQIWQQYFSAKDTVYAVIPK--EKFDLI 183
> cel:ZK945.3 puf-12; PUF (Pumilio/FBF) domain-containing family
member (puf-12); K14844 pumilio homology domain family member
6
Length=766
Score = 28.1 bits (61), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 24/39 (61%), Gaps = 3/39 (7%)
Query 7 KEKNAFLFICKF---LDDKRAKEVRELWEEYERGETPEA 42
KE+ AFL K + RA++ +ELWE+ G+TP+A
Sbjct 227 KERKAFLKELKLKRKPEGARAQKCKELWEKIRMGKTPKA 265
> cel:T21C9.3 hypothetical protein
Length=565
Score = 27.7 bits (60), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 5 REKEKNAFLFICKFLDDKRAKEVRELWEEY 34
R K AF+FI FL K+V +L+EEY
Sbjct 26 RRARKYAFVFIVAFLAVLTVKDVFDLFEEY 55
Lambda K H
0.319 0.131 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2057481480
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40