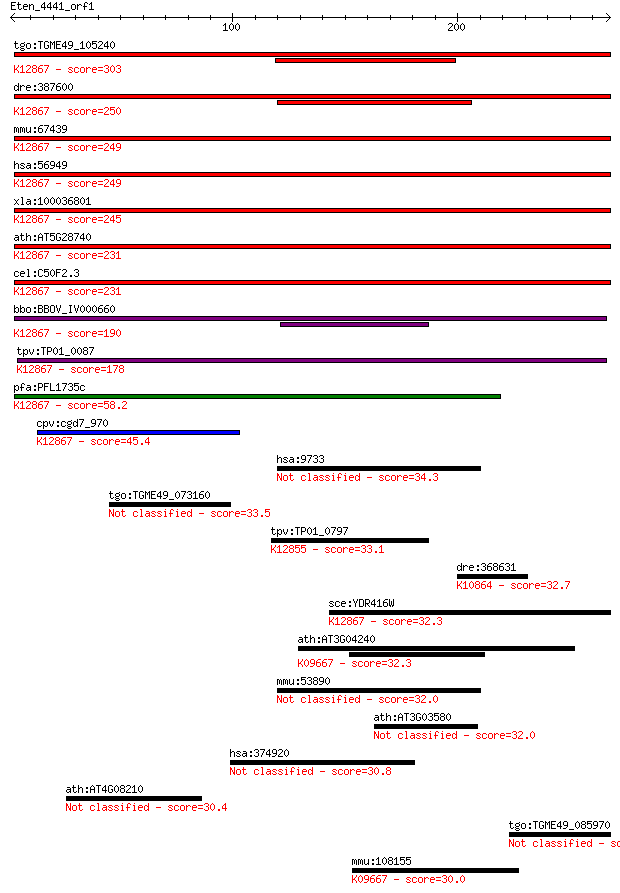
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4441_orf1
Length=267
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_105240 XPA-binding protein, putative ; K12867 pre-m... 303 5e-82
dre:387600 xab2, MGC198247, zgc:63498, zgc:63949; XPA binding ... 250 5e-66
mmu:67439 Xab2, 0610041O14Rik, AV025587; XPA binding protein 2... 249 8e-66
hsa:56949 XAB2, DKFZp762C1015, HCNP, HCRN, NTC90, SYF1; XPA bi... 249 9e-66
xla:100036801 xab2; XPA binding protein 2; K12867 pre-mRNA-spl... 245 1e-64
ath:AT5G28740 transcription-coupled DNA repair protein-related... 231 2e-60
cel:C50F2.3 hypothetical protein; K12867 pre-mRNA-splicing fac... 231 3e-60
bbo:BBOV_IV000660 21.m02991; XBA-binding protein 2; K12867 pre... 190 5e-48
tpv:TP01_0087 adapter protein; K12867 pre-mRNA-splicing factor... 178 2e-44
pfa:PFL1735c RNA-processing protein, putative; K12867 pre-mRNA... 58.2 3e-08
cpv:cgd7_970 Syf1p. protein with 8 HAT domains ; K12867 pre-mR... 45.4 2e-04
hsa:9733 SART3, DSAP1, KIAA0156, MGC138188, P100, RP11-13G14, ... 34.3 0.52
tgo:TGME49_073160 hypothetical protein 33.5 0.77
tpv:TP01_0797 hypothetical protein; K12855 pre-mRNA-processing... 33.1 0.98
dre:368631 pms1, PMSL1, si:dz164h20.2, si:dz72b14.2; PMS1 post... 32.7 1.4
sce:YDR416W SYF1, NTC90; Member of the NineTeen Complex (NTC) ... 32.3 1.9
ath:AT3G04240 SEC; SEC (secret agent); transferase, transferri... 32.3 2.1
mmu:53890 Sart3, AU045857, mKIAA0156; squamous cell carcinoma ... 32.0 2.4
ath:AT3G03580 pentatricopeptide (PPR) repeat-containing protein 32.0 2.6
hsa:374920 C19orf68, MGC50339; chromosome 19 open reading fram... 30.8 5.5
ath:AT4G08210 pentatricopeptide (PPR) repeat-containing protein 30.4 6.9
tgo:TGME49_085970 hypothetical protein 30.4 7.9
mmu:108155 Ogt, 1110038P24Rik, 4831420N21Rik, AI115525, Ogtl; ... 30.0 9.5
> tgo:TGME49_105240 XPA-binding protein, putative ; K12867 pre-mRNA-splicing
factor SYF1
Length=966
Score = 303 bits (775), Expect = 5e-82, Method: Compositional matrix adjust.
Identities = 150/289 (51%), Positives = 203/289 (70%), Gaps = 26/289 (8%)
Query 3 RCLVHLSRMPEIWRMNLKFLQQQQLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCG 62
R LVHLSRMP+IW + + FL++Q+LLTR RR FD AL++LAVTQH+ +W + +VK G
Sbjct 126 RALVHLSRMPKIWMLFVDFLKRQKLLTRTRRAFDRALQSLAVTQHDQVWDRYIQFVKEAG 185
Query 63 VPETAVCLYRRWIMIEPERTDEFALYLSS--IGRYDEAASHLAAVASDSTITTASGRTRH 120
V ET + +YRR +M+ PE+ ++F YL S +GRYD+AA LA V +D + T RT+H
Sbjct 186 VVETTLRVYRRCLMLLPEKVEDFIAYLQSPEVGRYDDAARLLAEVVNDESSETQ--RTKH 243
Query 121 ELWLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDI 180
ELWLELC LV HP I S+R E +LRSGI+RFSD V LWC+LASH+VR+G L K RD+
Sbjct 244 ELWLELCDLVCKHPREIKSLRAEAVLRSGISRFSDQVGKLWCALASHFVRLGQLEKTRDV 303
Query 181 YEEAVSSVSTIRDLAVVYEAYAAFEEAVVA---QLLQEQQQASPEAAAA----------- 226
+EEA+ V T+ DLA+VY+A+ FEE+++A + L++++ A P+ AA+
Sbjct 304 FEEALCGVGTLHDLALVYDAFVQFEESLLAAKMKELEDEENAGPDCAASEDAADRRERKR 363
Query 227 --------AAKDIDFAIARLERLTXRRPLLISSCKLRQNPHNVHEWLAR 267
++++DF + RLE LT RRPLL+SSCKLRQNPHNVHEWLAR
Sbjct 364 RRKEKKKQQSEEVDFLMTRLEFLTERRPLLVSSCKLRQNPHNVHEWLAR 412
Score = 34.3 bits (77), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 31/99 (31%), Positives = 49/99 (49%), Gaps = 21/99 (21%)
Query 119 RHELWLELCKLVAAHPESIHS--MRVEDILRSGIAR----FSDAVAS------------L 160
R L + CKL +P ++H RV D+ + A+ FS+AVA+ L
Sbjct 389 RRPLLVSSCKL-RQNPHNVHEWLARV-DLFKGDTAKEVETFSEAVATVDPQQAVGRVSVL 446
Query 161 WCSLASHYVRMGLLAKARDIYEEAVSS-VSTIRDLAVVY 198
W + A +Y G L AR I+E+A + V T+ +LA ++
Sbjct 447 WIAFARYYEDRGDLPNARLIFEKATKARVRTVDELASIW 485
> dre:387600 xab2, MGC198247, zgc:63498, zgc:63949; XPA binding
protein 2; K12867 pre-mRNA-splicing factor SYF1
Length=851
Score = 250 bits (638), Expect = 5e-66, Method: Compositional matrix adjust.
Identities = 115/265 (43%), Positives = 173/265 (65%), Gaps = 2/265 (0%)
Query 3 RCLVHLSRMPEIWRMNLKFLQQQQLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCG 62
R LV + +MP IW +F+ Q +TR RRTFD ALRAL +TQH +WP + + ++
Sbjct 96 RALVFMHKMPRIWIDYCQFMVSQCKITRSRRTFDRALRALPITQHPRIWPLYLRFARNLP 155
Query 63 VPETAVCLYRRWIMIEPERTDEFALYLSSIGRYDEAASHLAAVASDSTITTASGRTRHEL 122
+PETA+ +YRR++ + PE +E+ YL S+GR DEAA LAAV +D + G++ ++L
Sbjct 156 LPETAIRVYRRYLKLSPENAEEYIDYLRSVGRLDEAALRLAAVVNDENFVSKEGKSNYQL 215
Query 123 WLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDIYE 182
W ELC L++ +P+ + S+ V I+R G+ RF+D + LWCS+A +Y+R G KARD+YE
Sbjct 216 WHELCDLISQNPDKVTSLNVGAIIRGGLTRFTDQLGKLWCSMADYYIRSGHFEKARDVYE 275
Query 183 EAVSSVSTIRDLAVVYEAYAAFEEAVVAQLLQEQQQASPEAAAAAAKDIDFAIARLERLT 242
EA+ +V T+RD V+++YA FEE+++A ++ + + ++ +AR E L
Sbjct 276 EAILTVVTVRDFTQVFDSYAQFEESMIAAKMETTSELGQDEDDDIDLEL--RLARFESLI 333
Query 243 XRRPLLISSCKLRQNPHNVHEWLAR 267
RRPLL++S LRQNPHNVHEW R
Sbjct 334 TRRPLLLNSVLLRQNPHNVHEWHKR 358
Score = 33.9 bits (76), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 36/87 (41%), Gaps = 2/87 (2%)
Query 120 HELWLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARD 179
HE W + KL P I + E + + + SLW S A Y + AR
Sbjct 353 HE-WHKRVKLYEGQPRQIINTYTEAVQTIDPMKATGKPHSLWVSFAKFYEDNEQIDDART 411
Query 180 IYEEAVS-SVSTIRDLAVVYEAYAAFE 205
I+E+A + + DLA V+ Y E
Sbjct 412 IFEKATKVNYKQVDDLAAVWCEYGEME 438
> mmu:67439 Xab2, 0610041O14Rik, AV025587; XPA binding protein
2; K12867 pre-mRNA-splicing factor SYF1
Length=855
Score = 249 bits (636), Expect = 8e-66, Method: Compositional matrix adjust.
Identities = 115/265 (43%), Positives = 172/265 (64%), Gaps = 2/265 (0%)
Query 3 RCLVHLSRMPEIWRMNLKFLQQQQLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCG 62
R V + +MP +W +FL Q +T RRTFD ALRAL +TQH +WP + +++S
Sbjct 101 RAFVFMHKMPRLWLDYCQFLMDQGRVTHTRRTFDRALRALPITQHSRIWPLYLRFLRSHP 160
Query 63 VPETAVCLYRRWIMIEPERTDEFALYLSSIGRYDEAASHLAAVASDSTITTASGRTRHEL 122
+PETAV YRR++ + PE +E+ YL S R DEAA LA V +D + +G++ ++L
Sbjct 161 LPETAVRGYRRFLKLSPESAEEYIEYLKSSDRLDEAAQRLATVVNDERFVSKAGKSNYQL 220
Query 123 WLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDIYE 182
W ELC L++ +P+ + S+ V+ I+R G+ RF+D + LWCSLA +Y+R G KARD+YE
Sbjct 221 WHELCDLISQNPDKVQSLNVDAIIRGGLTRFTDQLGKLWCSLADYYIRSGHFEKARDVYE 280
Query 183 EAVSSVSTIRDLAVVYEAYAAFEEAVVAQLLQEQQQASPEAAAAAAKDIDFAIARLERLT 242
EA+ +V T+RD V+++YA FEE+++A ++ + E D++ +AR E+L
Sbjct 281 EAIRTVMTVRDFTQVFDSYAQFEESMIAAKMETASELGREEEDDV--DLELRLARFEQLI 338
Query 243 XRRPLLISSCKLRQNPHNVHEWLAR 267
RRPLL++S LRQNPH+VHEW R
Sbjct 339 SRRPLLLNSVLLRQNPHHVHEWHKR 363
> hsa:56949 XAB2, DKFZp762C1015, HCNP, HCRN, NTC90, SYF1; XPA
binding protein 2; K12867 pre-mRNA-splicing factor SYF1
Length=855
Score = 249 bits (635), Expect = 9e-66, Method: Compositional matrix adjust.
Identities = 115/265 (43%), Positives = 172/265 (64%), Gaps = 2/265 (0%)
Query 3 RCLVHLSRMPEIWRMNLKFLQQQQLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCG 62
R V + +MP +W +FL Q +T RRTFD ALRAL +TQH +WP + +++S
Sbjct 101 RAFVFMHKMPRLWLDYCQFLMDQGRVTHTRRTFDRALRALPITQHSRIWPLYLRFLRSHP 160
Query 63 VPETAVCLYRRWIMIEPERTDEFALYLSSIGRYDEAASHLAAVASDSTITTASGRTRHEL 122
+PETAV YRR++ + PE +E+ YL S R DEAA LA V +D + +G++ ++L
Sbjct 161 LPETAVRGYRRFLKLSPESAEEYIEYLKSSDRLDEAAQRLATVVNDERFVSKAGKSNYQL 220
Query 123 WLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDIYE 182
W ELC L++ +P+ + S+ V+ I+R G+ RF+D + LWCSLA +Y+R G KARD+YE
Sbjct 221 WHELCDLISQNPDKVQSLNVDAIIRGGLTRFTDQLGKLWCSLADYYIRSGHFEKARDVYE 280
Query 183 EAVSSVSTIRDLAVVYEAYAAFEEAVVAQLLQEQQQASPEAAAAAAKDIDFAIARLERLT 242
EA+ +V T+RD V+++YA FEE+++A ++ + E D++ +AR E+L
Sbjct 281 EAIRTVMTVRDFTQVFDSYAQFEESMIAAKMETASELGREEEDDV--DLELRLARFEQLI 338
Query 243 XRRPLLISSCKLRQNPHNVHEWLAR 267
RRPLL++S LRQNPH+VHEW R
Sbjct 339 SRRPLLLNSVLLRQNPHHVHEWHKR 363
> xla:100036801 xab2; XPA binding protein 2; K12867 pre-mRNA-splicing
factor SYF1
Length=838
Score = 245 bits (626), Expect = 1e-64, Method: Compositional matrix adjust.
Identities = 117/265 (44%), Positives = 171/265 (64%), Gaps = 2/265 (0%)
Query 3 RCLVHLSRMPEIWRMNLKFLQQQQLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCG 62
R LV + +MP IW +FL Q +TR RRTFD ALRAL +TQH +WP + +V++
Sbjct 92 RALVFMHKMPRIWLDYCQFLMDQCKITRARRTFDRALRALPITQHHRIWPLYLRFVRAHP 151
Query 63 VPETAVCLYRRWIMIEPERTDEFALYLSSIGRYDEAASHLAAVASDSTITTASGRTRHEL 122
+PETAV +YRR++ + PE +E+ YL SI R DEAAS LAA+ + + G++ ++L
Sbjct 152 LPETAVRVYRRYLKLSPENAEEYIEYLRSIDRLDEAASRLAAIVNQDGFVSKEGKSNYQL 211
Query 123 WLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDIYE 182
W +LC L++ HP S+ S+ I+R G+ RF+D LWC+LA ++ R G KARD+YE
Sbjct 212 WQQLCTLLSQHPGSVRSLDAAAIIRGGLTRFTDQRGKLWCALAEYHTRSGHFEKARDVYE 271
Query 183 EAVSSVSTIRDLAVVYEAYAAFEEAVVAQLLQEQQQASPEAAAAAAKDIDFAIARLERLT 242
EA+ +V+T+RD V+++YA FEE+V+A ++ E ++ +AR E+L
Sbjct 272 EAIQTVTTVRDFTQVFDSYAQFEESVIAAKMETVSDLGKEDEDDLDLEL--RLARFEQLI 329
Query 243 XRRPLLISSCKLRQNPHNVHEWLAR 267
RRPLL+++ LRQNPHNVHEW R
Sbjct 330 ERRPLLLNAVLLRQNPHNVHEWHKR 354
> ath:AT5G28740 transcription-coupled DNA repair protein-related;
K12867 pre-mRNA-splicing factor SYF1
Length=917
Score = 231 bits (589), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 120/299 (40%), Positives = 170/299 (56%), Gaps = 34/299 (11%)
Query 3 RCLVHLSRMPEIWRMNLKFLQQQQLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCG 62
R LV + +MP IW M L+ L QQL+TR RRTFD AL AL VTQH+ +W + +V G
Sbjct 93 RGLVTMHKMPRIWVMYLQTLTVQQLITRTRRTFDRALCALPVTQHDRIWEPYLVFVSQNG 152
Query 63 VP-ETAVCLYRRWIMIEPERTDEFALYLSSIGRYDEAASHLAAVASDSTITTASGRTRHE 121
+P ET++ +YRR++M +P +EF +L R+ E+A LA+V +D + G+T+H+
Sbjct 153 IPIETSLRVYRRYLMYDPSHIEEFIEFLVKSERWQESAERLASVLNDDKFYSIKGKTKHK 212
Query 122 LWLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDIY 181
LWLELC+L+ H I + V+ I+R GI +F+D V LW SLA +Y+R LL KARDIY
Sbjct 213 LWLELCELLVHHANVISGLNVDAIIRGGIRKFTDEVGMLWTSLADYYIRKNLLEKARDIY 272
Query 182 EEAVSSVSTIRDLAVVYEAYAAFEEAVVAQLLQEQQQASPEAAAAAA------------- 228
EE + V T+RD +V+++ Y+ FEE+ VA+ ++ + E
Sbjct 273 EEGMMKVVTVRDFSVIFDVYSRFEESTVAKKMEMMSSSDEEDENEENGVEDDEEDVRLNF 332
Query 229 --------------------KDIDFAIARLERLTXRRPLLISSCKLRQNPHNVHEWLAR 267
D+D +ARLE L RRP L +S LRQNPHNV +W R
Sbjct 333 NLSVKELQRKILNGFWLNDDNDVDLRLARLEELMNRRPALANSVLLRQNPHNVEQWHRR 391
> cel:C50F2.3 hypothetical protein; K12867 pre-mRNA-splicing factor
SYF1
Length=855
Score = 231 bits (588), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 112/266 (42%), Positives = 167/266 (62%), Gaps = 5/266 (1%)
Query 3 RCLVHLSRMPEIWRMNLKFLQQQQLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCG 62
RCL+ L +MP IW + + ++ L+T RR FD ALR+L VTQH +W + ++ S
Sbjct 108 RCLMRLHKMPRIWICYCEVMIKRGLITETRRVFDRALRSLPVTQHMRIWTLYIGFLTSHD 167
Query 63 VPETAVCLYRRWIMIEPERTDEFALYLSSIGRYDEAASHLAAVASDSTITTASGRTRHEL 122
+PET + +YRR++ + P+ +++ YL + DEAA L + + + GRT H+L
Sbjct 168 LPETTIRVYRRYLKMNPKAREDYVEYLIERDQIDEAAKELTTLVNQDQNVSEKGRTAHQL 227
Query 123 WLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDIYE 182
W +LC L++ +P I S+ V+ I+R GI R++D V LWCSLA +Y+R +ARD+YE
Sbjct 228 WTQLCDLISKNPVKIFSLNVDAIIRQGIYRYTDQVGFLWCSLADYYIRSAEFERARDVYE 287
Query 183 EAVSSVSTIRDLAVVYEAYAAFEEAVVAQLLQE-QQQASPEAAAAAAKDIDFAIARLERL 241
EA++ VST+RD A VY+AYAAFEE V+ ++QE +Q PE D+++ R + L
Sbjct 288 EAIAKVSTVRDFAQVYDAYAAFEEREVSIMMQEVEQSGDPEEEV----DLEWMFQRYQHL 343
Query 242 TXRRPLLISSCKLRQNPHNVHEWLAR 267
R+ L++S LRQNPHNV EWL R
Sbjct 344 MERKNELMNSVLLRQNPHNVGEWLNR 369
> bbo:BBOV_IV000660 21.m02991; XBA-binding protein 2; K12867 pre-mRNA-splicing
factor SYF1
Length=796
Score = 190 bits (482), Expect = 5e-48, Method: Compositional matrix adjust.
Identities = 91/263 (34%), Positives = 153/263 (58%), Gaps = 9/263 (3%)
Query 3 RCLVHLSRMPEIWRMNLKFLQQQQLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCG 62
RC++H+ P I+ + +FL+ Q +++R RRT+D AL L +TQH ++W + +++VK
Sbjct 151 RCIIHVYAAPAIYILYGQFLRTQNMISRTRRTYDRALLNLPITQHMMIWQQYIEFVKEVD 210
Query 63 VPETAVCLYRRWIMIEPERTDEFALYLSSIGRYDEAASHLAAVASDSTITTASGRTRHEL 122
+ + +R+I ++P + L YDEA L + +D + SG+T+++L
Sbjct 211 LLPMGKAVLKRYIQLQPNTRESLYKMLKQHEHYDEACIVLCELLNDGKFVSESGKTQYDL 270
Query 123 WLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDIYE 182
W+ELC+L+ + + I S+ +E I++ GIA++SD VA LW LA Y+ G + ARD+YE
Sbjct 271 WVELCELIRDYSQYIRSVPIEAIIKEGIAKYSDQVAQLWIILADIYILRGQMLNARDVYE 330
Query 183 EAVSSVSTIRDLAVVYEAYAAFEEAVVAQLLQEQQQASPEAAAAAAKDIDFAIARLERLT 242
EA+ SV+T++D + +++ YA F E Q + A D+ + RLE L
Sbjct 331 EALKSVTTVQDFSTIFDVYAKFLEKYAKQ---------RKKLRGADLDVVMTVDRLENLI 381
Query 243 XRRPLLISSCKLRQNPHNVHEWL 265
R +L++ KL+QN HNV+ WL
Sbjct 382 NTRAMLMAKVKLKQNAHNVYNWL 404
Score = 32.0 bits (71), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 31/66 (46%), Gaps = 0/66 (0%)
Query 121 ELWLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDI 180
ELW H + + R+ + G +F D +AS+WC+ ++R L +A ++
Sbjct 439 ELWTSYASYFENHVDVDAADRIFEKAVEGNYKFVDDLASVWCAWVEMHIRHNNLKRALEL 498
Query 181 YEEAVS 186
+AV
Sbjct 499 SRQAVD 504
> tpv:TP01_0087 adapter protein; K12867 pre-mRNA-splicing factor
SYF1
Length=839
Score = 178 bits (451), Expect = 2e-44, Method: Composition-based stats.
Identities = 92/274 (33%), Positives = 153/274 (55%), Gaps = 20/274 (7%)
Query 4 CLVHLSRMPEIWRMNLKFLQQQQLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCGV 63
CL+H P + + FL+ Q +T+VRR +D AL +A+TQH L+W E + +V +
Sbjct 172 CLIHNFAYPTFYLLYGAFLRFQHRITKVRRLYDKALLNIAITQHHLIWDEYLRFVNEVDL 231
Query 64 PETAVCLYRRWIMIEPERTDEFALYLSSIGRYDEAASHLAAVASDSTITTASGRTRHELW 123
+YRR+I ++P + +L G YD+AA L + +D T + SG++ ++LW
Sbjct 232 LPLGKAVYRRYIQLKPSYREVLYEFLKRHGSYDDAAQVLYKLLNDHTFASESGKSSYDLW 291
Query 124 LELCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDIYEE 183
+ELC+L+ H ++I S+ VE +++ GI +++D VA+LW LA Y+ G L ARD YEE
Sbjct 292 IELCELIRDHSDAITSIPVETLIKEGIGKYTDQVATLWIILADIYIVRGQLNIARDTYEE 351
Query 184 AVSSVSTIRDLAVVYEAYAAFEEAVVAQLLQEQQQASPEAAAAAAKDIDF---------- 233
A+ V+T++D +++++ YA F L+ + S + ++
Sbjct 352 ALDRVTTVQDFSIIFDVYAKF--------LENYAKQSNKLGYVLINNLHLHYIRTDHLET 403
Query 234 --AIARLERLTXRRPLLISSCKLRQNPHNVHEWL 265
+ RLE L R LL++S KL+QN HNV+ W+
Sbjct 404 LMTVERLESLVNNRALLLASVKLKQNIHNVYNWI 437
> pfa:PFL1735c RNA-processing protein, putative; K12867 pre-mRNA-splicing
factor SYF1
Length=1031
Score = 58.2 bits (139), Expect = 3e-08, Method: Composition-based stats.
Identities = 43/222 (19%), Positives = 104/222 (46%), Gaps = 12/222 (5%)
Query 3 RCLVHLSRMPEIWRMNLKFLQQQQLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCG 62
+CL+++ I+ M ++FL Q+ + ++R+ F+ +L+ + + Q LW + + +
Sbjct 240 QCLLYMYNFKSIYIMYIQFLYIQRDVKKIRQIFNLSLQNVYLNQQNDLWECQLLFNEKIN 299
Query 63 VPETAVCLYRRWIMIEPERTDEFALYLSSIGRYDEAASHLAAVASDSTITTASGRTRHEL 122
+R++ I PE+ + Y A + + +++++
Sbjct 300 NKVINYEYIKRYVTIYPEQIIHLFKHYVKYKMYKNAMITFFYILNSEDNFDLGNFSKYDI 359
Query 123 WLELCKLV----AAHPESIHSMRVEDILRSGIARFS--DAVASLWCSLASHYVRMGLLAK 176
+ E+ KL+ + + + IH +LR+ + F +++ S++ LA++++ G K
Sbjct 360 YQEIYKLLNSKGSLNNDIIH------LLRNNLYIFKNYESITSIYILLANNFIYDGRWNK 413
Query 177 ARDIYEEAVSSVSTIRDLAVVYEAYAAFEEAVVAQLLQEQQQ 218
A D YEE +S T+ D +++ Y + ++ + EQ++
Sbjct 414 AMDSYEEGISECYTVNDFITLFDNYIEMLKMLIDLNIYEQEE 455
> cpv:cgd7_970 Syf1p. protein with 8 HAT domains ; K12867 pre-mRNA-splicing
factor SYF1
Length=1020
Score = 45.4 bits (106), Expect = 2e-04, Method: Composition-based stats.
Identities = 23/94 (24%), Positives = 48/94 (51%), Gaps = 6/94 (6%)
Query 13 EIWRMNLKFLQQQQL-LTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCGVPETAVCLY 71
++W +L++ +L T R D +L++L + QH +W ++Y +PE ++ +
Sbjct 171 DLWLRYNAYLRKSRLEFTNSRLVLDRSLKSLPIEQHHKIWERYLEYSMEMNIPELSISIS 230
Query 72 RRWIMIEPERTDEFALYLSSI---GRYDEAASHL 102
RR+I+ + +Y+ ++ GRY+E L
Sbjct 231 RRFILF--SYVEGIRMYIQALIDGGRYEECLDKL 262
> hsa:9733 SART3, DSAP1, KIAA0156, MGC138188, P100, RP11-13G14,
TIP110, p110, p110(nrb); squamous cell carcinoma antigen recognized
by T cells 3
Length=963
Score = 34.3 bits (77), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 46/97 (47%), Gaps = 10/97 (10%)
Query 120 HELWLE-LCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYV----RMGLL 174
ELWLE L ++ + + V D+ + + ++W + V + G L
Sbjct 146 EELWLEWLHDEISMAQDGLDREHVYDLFEKAVKDY--ICPNIWLEYGQYSVGGIGQKGGL 203
Query 175 AKARDIYEEAVSSVS--TIRDLAVVYEAYAAFEEAVV 209
K R ++E A+SSV + LA ++EAY FE A+V
Sbjct 204 EKVRSVFERALSSVGLHMTKGLA-LWEAYREFESAIV 239
> tgo:TGME49_073160 hypothetical protein
Length=243
Score = 33.5 bits (75), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 26/54 (48%), Gaps = 0/54 (0%)
Query 45 TQHELLWPEMMDYVKSCGVPETAVCLYRRWIMIEPERTDEFALYLSSIGRYDEA 98
++ +LL+ E D G PE C+ + +PE E A +L GR +EA
Sbjct 105 SERQLLFAEAADGDARAGKPEEESCMDASGLAGDPEPAGERACFLEPQGRLEEA 158
> tpv:TP01_0797 hypothetical protein; K12855 pre-mRNA-processing
factor 6
Length=1032
Score = 33.1 bits (74), Expect = 0.98, Method: Composition-based stats.
Identities = 24/72 (33%), Positives = 37/72 (51%), Gaps = 8/72 (11%)
Query 117 RTRHELWLELCKLVAAH--PESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLL 174
+TR LWL L +L + H P+ +VED LRS ++ ++ LW A H G +
Sbjct 576 KTRSSLWLALVELESKHGTPD-----KVEDHLRSAVSYCPNS-EILWLMYAKHKWVEGDV 629
Query 175 AKARDIYEEAVS 186
+RDI A++
Sbjct 630 ESSRDILSRALT 641
> dre:368631 pms1, PMSL1, si:dz164h20.2, si:dz72b14.2; PMS1 postmeiotic
segregation increased 1 (S. cerevisiae); K10864 DNA
mismatch repair protein PMS1
Length=896
Score = 32.7 bits (73), Expect = 1.4, Method: Composition-based stats.
Identities = 14/31 (45%), Positives = 22/31 (70%), Gaps = 0/31 (0%)
Query 200 AYAAFEEAVVAQLLQEQQQASPEAAAAAAKD 230
A++ FE+ +Q+LQE +ASP+ AAAK+
Sbjct 532 AFSLFEQDTRSQVLQENPKASPQDVTAAAKE 562
> sce:YDR416W SYF1, NTC90; Member of the NineTeen Complex (NTC)
that contains Prp19p and stabilizes U6 snRNA in catalytic
forms of the spliceosome containing U2, U5, and U6 snRNAs; null
mutant has splicing defect and arrests in G2/M; homologs
in human and C. elegans; K12867 pre-mRNA-splicing factor SYF1
Length=859
Score = 32.3 bits (72), Expect = 1.9, Method: Composition-based stats.
Identities = 26/127 (20%), Positives = 52/127 (40%), Gaps = 2/127 (1%)
Query 143 EDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDIYEEAVSSVSTIRDLAVVYEAYA 202
E+ +R + D L SLA +Y+ G L D+ ++++ D +Y Y
Sbjct 288 EEFMRQMNGIYPDKWLFLILSLAKYYISRGRLDSCGDLLKKSLQQTLRYSDFDRIYNFYL 347
Query 203 AFEEAVVAQLLQEQQQASPE--AAAAAAKDIDFAIARLERLTXRRPLLISSCKLRQNPHN 260
FE+ +L + ++ + + + +A E L + ++ LRQ+ +
Sbjct 348 LFEQECSQFILGKLKENDSKFFNQKDWTEKLQAHMATFESLINLYDIYLNDVALRQDSNL 407
Query 261 VHEWLAR 267
V W+ R
Sbjct 408 VETWMKR 414
> ath:AT3G04240 SEC; SEC (secret agent); transferase, transferring
glycosyl groups; K09667 polypeptide N-acetylglucosaminyltransferase
[EC:2.4.1.-]
Length=977
Score = 32.3 bits (72), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 35/142 (24%), Positives = 57/142 (40%), Gaps = 21/142 (14%)
Query 129 LVAAHPESIHSMRVEDILRSGIARFSDAV------ASLWCSLASHYVRMGLLAKARDIYE 182
LV AH + M+ + ++ + + +AV A W +LA ++ G L +A Y+
Sbjct 188 LVDAHSNLGNLMKAQGLIHEAYSCYLEAVRIQPTFAIAWSNLAGLFMESGDLNRALQYYK 247
Query 183 EAVSSVSTIRD----LAVVYEAYAAFEEAVVAQLLQEQQQASPEAAAAAAK--------- 229
EAV D L VY+A EA++ Q Q P +A A
Sbjct 248 EAVKLKPAFPDAYLNLGNVYKALGRPTEAIMC--YQHALQMRPNSAMAFGNIASIYYEQG 305
Query 230 DIDFAIARLERLTXRRPLLISS 251
+D AI ++ R P + +
Sbjct 306 QLDLAIRHYKQALSRDPRFLEA 327
Score = 30.4 bits (67), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 39/71 (54%), Gaps = 15/71 (21%)
Query 152 RFSDAVASLWCSLASHYVRMGLLAKARDIYEEAVS-------SVSTIRDL----AVVYEA 200
F+DA W +LAS Y+R G L++A ++A+S + S + +L +++EA
Sbjct 153 NFADA----WSNLASAYMRKGRLSEATQCCQQALSLNPLLVDAHSNLGNLMKAQGLIHEA 208
Query 201 YAAFEEAVVAQ 211
Y+ + EAV Q
Sbjct 209 YSCYLEAVRIQ 219
> mmu:53890 Sart3, AU045857, mKIAA0156; squamous cell carcinoma
antigen recognized by T-cells 3
Length=962
Score = 32.0 bits (71), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 46/97 (47%), Gaps = 10/97 (10%)
Query 120 HELWLE-LCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYV----RMGLL 174
ELWLE L ++ + + V ++ + + ++W + V + G L
Sbjct 147 EELWLEWLHDEISMAMDGLDREHVYELFERAVKDY--ICPNIWLEYGQYSVGGIGQKGGL 204
Query 175 AKARDIYEEAVSSVS--TIRDLAVVYEAYAAFEEAVV 209
K R ++E A+SSV + LA ++EAY FE A+V
Sbjct 205 EKVRSVFERALSSVGLHMTKGLA-IWEAYREFESAIV 240
> ath:AT3G03580 pentatricopeptide (PPR) repeat-containing protein
Length=882
Score = 32.0 bits (71), Expect = 2.6, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 27/47 (57%), Gaps = 1/47 (2%)
Query 163 SLASHYVRMGLLAKARDIYEE-AVSSVSTIRDLAVVYEAYAAFEEAV 208
+L Y RMGLL +AR +++E V + + L Y ++ +EEA+
Sbjct 146 ALVDMYSRMGLLTRARQVFDEMPVRDLVSWNSLISGYSSHGYYEEAL 192
> hsa:374920 C19orf68, MGC50339; chromosome 19 open reading frame
68
Length=578
Score = 30.8 bits (68), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 32/82 (39%), Gaps = 0/82 (0%)
Query 99 ASHLAAVASDSTITTASGRTRHELWLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVA 158
A L + S + +GR LW LC+L A + + + ++ G A F D
Sbjct 269 AQGLETLFSKAQELGGAGREDPGLWSRLCRLAGASSPAAYDEALAELHAHGPAAFVDYFE 328
Query 159 SLWCSLASHYVRMGLLAKARDI 180
W +VR ARD+
Sbjct 329 RNWEPRRDMWVRFRAFEAARDL 350
> ath:AT4G08210 pentatricopeptide (PPR) repeat-containing protein
Length=686
Score = 30.4 bits (67), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 27/60 (45%), Gaps = 5/60 (8%)
Query 26 QLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCGVPETAVCLYRRWIMIEPERTDEF 85
+LL+ + FD VT W M+ S G P A+ LYRR + E E +EF
Sbjct 54 RLLSDAHKVFDEMSERNIVT-----WTTMVSGYTSDGKPNKAIELYRRMLDSEEEAANEF 108
> tgo:TGME49_085970 hypothetical protein
Length=912
Score = 30.4 bits (67), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 25/56 (44%), Gaps = 11/56 (19%)
Query 223 AAAAAAKDIDFAIARLERLTXRRPL-----------LISSCKLRQNPHNVHEWLAR 267
AA + A+D++F RL P L S KLR++PH H+W R
Sbjct 280 AANSVAEDLEFLCRRLSVPNLPHPTHPFRLKLLLDALFESFKLRKDPHYAHDWAQR 335
> mmu:108155 Ogt, 1110038P24Rik, 4831420N21Rik, AI115525, Ogtl;
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl
transferase);
K09667 polypeptide N-acetylglucosaminyltransferase [EC:2.4.1.-]
Length=1046
Score = 30.0 bits (66), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 39/78 (50%), Gaps = 10/78 (12%)
Query 153 FSDAVASLWCSLASHYVRMGLLAKARDIYEEAV----SSVSTIRDLAVVYEAYAAFEEAV 208
F DA +C+LA+ G +A+A D Y A+ + ++ +LA + EEAV
Sbjct 292 FPDA----YCNLANALKEKGSVAEAEDCYNTALRLCPTHADSLNNLANIKREQGNIEEAV 347
Query 209 VAQLLQEQQQASPEAAAA 226
+L ++ + PE AAA
Sbjct 348 --RLYRKALEVFPEFAAA 363
Lambda K H
0.323 0.131 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9954723792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40