bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4458_orf1
Length=217
Score E
Sequences producing significant alignments: (Bits) Value
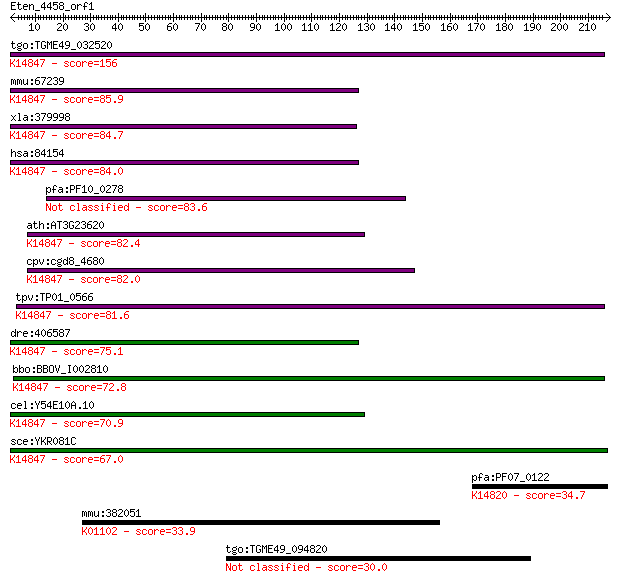
tgo:TGME49_032520 brix domain-containing protein ; K14847 ribo... 156 6e-38
mmu:67239 Rpf2, 2810470K21Rik, AU040229, AU043242, Bxdc1; ribo... 85.9 9e-17
xla:379998 rpf2, MGC53329, bxdc1; ribosome production factor 2... 84.7 2e-16
hsa:84154 RPF2, BXDC1, FLJ21087, bA397G5.4; ribosome productio... 84.0 4e-16
pfa:PF10_0278 nucleolar preribosomal assembly protein, putative 83.6 5e-16
ath:AT3G23620 brix domain-containing protein; K14847 ribosome ... 82.4 1e-15
cpv:cgd8_4680 conserved protein; Py hit 23481194 ; K14847 ribo... 82.0 1e-15
tpv:TP01_0566 hypothetical protein; K14847 ribosome production... 81.6 2e-15
dre:406587 rpf2, bxdc1, ik:tdsubc_1a3, wu:fe17d08, wu:fj16d06,... 75.1 2e-13
bbo:BBOV_I002810 19.m02110; Brix domain containing protein; K1... 72.8 1e-12
cel:Y54E10A.10 hypothetical protein; K14847 ribosome productio... 70.9 3e-12
sce:YKR081C RPF2; Essential protein involved in the processing... 67.0 4e-11
pfa:PF07_0122 nucleolus BRIX protein, putative; K14820 ribosom... 34.7 0.26
mmu:382051 Pdp2, 4833426J09Rik, Gm1705, KIAA1348, mKIAA1348; p... 33.9 0.44
tgo:TGME49_094820 type I fatty acid synthase, putative (EC:6.2... 30.0 6.2
> tgo:TGME49_032520 brix domain-containing protein ; K14847 ribosome
production factor 2
Length=401
Score = 156 bits (394), Expect = 6e-38, Method: Compositional matrix adjust.
Identities = 98/231 (42%), Positives = 136/231 (58%), Gaps = 22/231 (9%)
Query 1 FENAEPIEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVAHYTPAAAFA 60
FE+ EYL +KN CG F SSSKKRP RLV GR++ N++LDM EF V HY A++F
Sbjct 79 FEDVSTAEYLCQKNDCGTFCMGSSSKKRPNRLVFGRMFANELLDMYEFQVVHYMGASSFT 138
Query 61 AVQAPAAGSAPLLLLQGGLWDVSDEMRNARNIFGDLFR-----AAEVPPDGPTQKLYLGG 115
V+ P GS PL+LLQG ++ ++ R RNI D+FR A V +G + KLYL G
Sbjct 139 GVEKPRLGSRPLVLLQGAAFESTEVHRGLRNIMADVFRGGASHAGAVAGEG-SAKLYLKG 197
Query 116 VDRLIAVSAVQATPGSAATAATT----AAAAKAVIGGSEDCETEA--PASGATTD-GTDA 168
+DR++A++A++ T G+A+ + A A I G+ + A P A D G
Sbjct 198 IDRVVAITALE-TDGAASHLSRKLHLEAGGTTATIEGAANAAVAATLPVPDAGNDQGGSG 256
Query 169 PAAD--SGYSGDVLICVRHYRIALVKTADAGGVGG--PAVKLVEVGPQIDL 215
P+AD + +G LIC+R YR+ + + G VG P V+LVEVGPQID+
Sbjct 257 PSADKATAAAGKPLICLRQYRLVMHR----GEVGSKVPRVELVEVGPQIDM 303
> mmu:67239 Rpf2, 2810470K21Rik, AU040229, AU043242, Bxdc1; ribosome
production factor 2 homolog (S. cerevisiae); K14847 ribosome
production factor 2
Length=273
Score = 85.9 bits (211), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 40/127 (31%), Positives = 70/127 (55%), Gaps = 9/127 (7%)
Query 1 FENAEPIEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVAHYTPAAAFA 60
FE+ +E+ ++K+ C LF + S +KKRP LV+GR+YD +LDM E + +
Sbjct 39 FEDQTSLEFFSKKSDCSLFMFGSHNKKRPNNLVIGRMYDYHVLDMIELGIEKFVSLKDIK 98
Query 61 AVQAPAAGSAPLLLLQGGLWDVSDEMRNARNIFGDLFRAAEVPPDGPT-QKLYLGGVDRL 119
+ P G+ P+L+ G +DV+++ R +N+ D FR GPT + L G++ +
Sbjct 99 TSKCP-EGTKPMLIFAGDDFDVTEDFRRLKNLLIDFFR-------GPTVSNVRLAGLEYV 150
Query 120 IAVSAVQ 126
+ +A+
Sbjct 151 LHFTALN 157
> xla:379998 rpf2, MGC53329, bxdc1; ribosome production factor
2 homolog; K14847 ribosome production factor 2
Length=308
Score = 84.7 bits (208), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 42/126 (33%), Positives = 69/126 (54%), Gaps = 9/126 (7%)
Query 1 FENAEPIEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVAHYTPAAAFA 60
FE+ +E+ A+K+ C LF + S +KKRP L+ GR+YD +LDM E V Y
Sbjct 72 FEDQTSLEFFAKKSDCSLFVFGSHNKKRPNDLIFGRMYDFHVLDMVELGVEKYVSLQEIK 131
Query 61 AVQAPAAGSAPLLLLQGGLWDVSDEMRNARNIFGDLFRAAEVPPDGPTQK-LYLGGVDRL 119
+ P G+ PLL+ G +++S++ R +++ D FR GPT + L G++ +
Sbjct 132 NCKCP-EGTKPLLIFAGDAFEMSEDYRRLKSLLIDFFR-------GPTVTGVRLAGLEHV 183
Query 120 IAVSAV 125
+ +AV
Sbjct 184 LHFTAV 189
> hsa:84154 RPF2, BXDC1, FLJ21087, bA397G5.4; ribosome production
factor 2 homolog (S. cerevisiae); K14847 ribosome production
factor 2
Length=306
Score = 84.0 bits (206), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 39/127 (30%), Positives = 71/127 (55%), Gaps = 9/127 (7%)
Query 1 FENAEPIEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVAHYTPAAAFA 60
FE+ +E+ ++K+ C LF + S +KKRP LV+GR+YD +LDM E + ++
Sbjct 72 FEDQTSLEFFSKKSDCSLFMFGSHNKKRPNNLVIGRMYDYHVLDMIELGIENFVSLKDIK 131
Query 61 AVQAPAAGSAPLLLLQGGLWDVSDEMRNARNIFGDLFRAAEVPPDGPT-QKLYLGGVDRL 119
+ P G+ P+L+ G +DV+++ R +++ D FR GPT + L G++ +
Sbjct 132 NSKCP-EGTKPMLIFAGDDFDVTEDYRRLKSLLIDFFR-------GPTVSNIRLAGLEYV 183
Query 120 IAVSAVQ 126
+ +A+
Sbjct 184 LHFTALN 190
> pfa:PF10_0278 nucleolar preribosomal assembly protein, putative
Length=483
Score = 83.6 bits (205), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 40/130 (30%), Positives = 65/130 (50%), Gaps = 6/130 (4%)
Query 14 NGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVAHYTPAAAFAAVQAPAAGSAPLL 73
N C F S+KK+P+R ++GR+Y+N+ILD F++ Y P F + + P++
Sbjct 95 NNCSFFFSVFSTKKKPSRFIIGRLYNNKILDYYVFSLISYIPLKLFPLSKEILYDTKPIV 154
Query 74 LLQGGLWDVSDEMRNARNIFGDLFRAAEVPPDGPTQKLYLGGVDRLIAVSAVQATPGSAA 133
L+QG ++ ++ R +NI D F+ V D ++K + RLI +SA Q A
Sbjct 155 LIQGSYFEQNETNRYVKNILFDFFKHKNV--DTFSKK----SIQRLIVISAYQKNNEINA 208
Query 134 TAATTAAAAK 143
A K
Sbjct 209 DLGKKVEAGK 218
> ath:AT3G23620 brix domain-containing protein; K14847 ribosome
production factor 2
Length=314
Score = 82.4 bits (202), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 41/124 (33%), Positives = 67/124 (54%), Gaps = 8/124 (6%)
Query 7 IEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVAHYTPAAAFAAVQ--A 64
+E+ ++K C +F Y S +KKRP LVLGR+YD+Q+ D+ E + ++ AF+ + A
Sbjct 78 LEFFSQKTDCSIFVYGSHTKKRPDNLVLGRMYDHQVYDLIEVGIENFKSLRAFSYDKKFA 137
Query 65 PAAGSAPLLLLQGGLWDVSDEMRNARNIFGDLFRAAEVPPDGPTQKLYLGGVDRLIAVSA 124
P G+ P + G ++ E+++ + + DLFR V L L G+DR SA
Sbjct 138 PHEGTKPFICFIGEGFENVSELKHLKEVLTDLFRGEVV------DNLNLTGLDRAYVCSA 191
Query 125 VQAT 128
+ T
Sbjct 192 ISPT 195
> cpv:cgd8_4680 conserved protein; Py hit 23481194 ; K14847 ribosome
production factor 2
Length=332
Score = 82.0 bits (201), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 47/143 (32%), Positives = 73/143 (51%), Gaps = 12/143 (8%)
Query 7 IEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVAHYTPAAAFAAVQAPA 66
+E L+ K+G + +A+SSKK P RL+ R YD+ ILDM EF V +Y + ++ P
Sbjct 81 MEQLSYKHGSSISIFANSSKKHPFRLIFTRYYDSHILDMYEFNVLNYKGIS--PNMELPK 138
Query 67 AGSAPLLLLQGGLWDVSDEMRNARNIFGDLFRAAEVPPDGPT---QKLYLGGVDRLIAVS 123
GS P+++ QG ++ D ++ R +F D F GP KL+L G D LI V+
Sbjct 139 YGSKPIVICQGAPFESDDVYKSIRTMFFDTF-------SGPIVRGSKLFLKGFDHLILVT 191
Query 124 AVQATPGSAATAATTAAAAKAVI 146
A + +T + + I
Sbjct 192 AYETDNEEINKQSTIIGSMNSKI 214
> tpv:TP01_0566 hypothetical protein; K14847 ribosome production
factor 2
Length=342
Score = 81.6 bits (200), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 62/219 (28%), Positives = 90/219 (41%), Gaps = 35/219 (15%)
Query 3 NAEPIEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVAHYTPAAAFAAV 62
N E + K G GL+ ASS+KKRP + GR+Y+ +ILD+ + V Y P F +
Sbjct 84 NHRECELICGKLGFGLYVVASSNKKRPISIGFGRMYNRKILDLCQLKVLQYKPTQHFNQL 143
Query 63 QAPAAGSAPLLLLQG-GLWDVSDEMRNARNIFGDLFRAAEVPPDGPTQ-KLYLGGVDRLI 120
S PL+++QG G + RNI D+F+ GPT K+ L + LI
Sbjct 144 DV-ELNSRPLIVVQGSGFGQQKGHLTTVRNILLDIFK-------GPTHTKISLDNIKHLI 195
Query 121 AVSAVQATPGSAATAATTAAAAKAVIGGSEDCETEAPASGATTDGTDAPAADSGYSGDVL 180
++ + + S+ T TT + E E E Y
Sbjct 196 TITLIDQS-NSSNTVDTTNT-----MDNEESVEVERRKV---------------YKNGPK 234
Query 181 ICVRHYRIALVK----TADAGGVGGPAVKLVEVGPQIDL 215
+ R Y I L+K + P V L E+GP ID
Sbjct 235 VLFRRYLITLLKPKKLEHNHSNTMLPRVSLSEIGPSIDF 273
> dre:406587 rpf2, bxdc1, ik:tdsubc_1a3, wu:fe17d08, wu:fj16d06,
xx:tdsubc_1a3, zgc:77150; ribosome production factor 2 homolog
(S. cerevisiae); K14847 ribosome production factor 2
Length=308
Score = 75.1 bits (183), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 33/126 (26%), Positives = 67/126 (53%), Gaps = 7/126 (5%)
Query 1 FENAEPIEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVAHYTPAAAFA 60
FE++ +E+ ++K+ C LF + S +KKRP L+ GR++D +LDM E + ++
Sbjct 72 FEDSTALEFFSKKSDCSLFLFGSHNKKRPNNLIFGRMFDFHVLDMFELGIENFRSLTDIK 131
Query 61 AVQAPAAGSAPLLLLQGGLWDVSDEMRNARNIFGDLFRAAEVPPDGPTQKLYLGGVDRLI 120
+ P G+ P+L+ G L++ E + +++ D F V + L G++ ++
Sbjct 132 MDKCP-EGTKPMLVFAGDLFETDREYQRLKSVLTDFFCGPRVSA------VRLAGLEHVL 184
Query 121 AVSAVQ 126
+A++
Sbjct 185 HFTALE 190
> bbo:BBOV_I002810 19.m02110; Brix domain containing protein;
K14847 ribosome production factor 2
Length=322
Score = 72.8 bits (177), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 61/217 (28%), Positives = 91/217 (41%), Gaps = 53/217 (24%)
Query 2 ENAEPIEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVAHYTPAAAFAA 61
N E ++ + GCGL+ ASS+KKRP L GR+YD +ILD Y P F
Sbjct 76 HNHRIAEMVSNRMGCGLYVVASSNKKRPLSLGFGRLYDGKILDCCHLRALKYVPFDFFKG 135
Query 62 VQ--APAAGSAPLLLLQGGLW-DVSDEMRNARNIFGDLFRAAEVPPDGPTQKLYLGGVDR 118
+ S PL++ QG + + S MR R+I D FR PP T + L +++
Sbjct 136 PNDLEISHNSYPLVVAQGAHFGEESGRMRTVRDILLDFFR----PPQ--TGCVSLENINQ 189
Query 119 LIAVSAVQATPGSAATAATTAAAAKAVIGGSEDCETEAPASGATTDGTDAPAADSGYSGD 178
++ +++V D E + P +
Sbjct 190 VLVLTSV-------------------------DGEGDQPGAQPQR--------------- 209
Query 179 VLICVRHYRIALVKTADAGGVGGPAVKLVEVGPQIDL 215
I +R Y+I L K++D + P V L E+GP IDL
Sbjct 210 --ILLRRYQINLNKSSDDPKL--PHVSLTEIGPHIDL 242
> cel:Y54E10A.10 hypothetical protein; K14847 ribosome production
factor 2
Length=297
Score = 70.9 bits (172), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 41/128 (32%), Positives = 64/128 (50%), Gaps = 7/128 (5%)
Query 1 FENAEPIEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVAHYTPAAAFA 60
FE+ PI K LF S+SKK+P L GR YD Q+LDM E + Y ++ F
Sbjct 69 FEDETPIVRAGSKFDTSLFVLGSNSKKKPNCLTFGRTYDGQLLDMAELRITSYKSSSNFE 128
Query 61 AVQAPAAGSAPLLLLQGGLWDVSDEMRNARNIFGDLFRAAEVPPDGPTQKLYLGGVDRLI 120
A + GS P ++L+G ++ +M+ N+ D FR +V + L G++ +I
Sbjct 129 AAKM-TLGSKPCVILEGAAFESDGDMKRIGNLMVDWFRGPKVDT------VRLEGLETVI 181
Query 121 AVSAVQAT 128
+A+ T
Sbjct 182 VFTALDET 189
> sce:YKR081C RPF2; Essential protein involved in the processing
of pre-rRNA and the assembly of the 60S ribosomal subunit;
interacts with ribosomal protein L11; localizes predominantly
to the nucleolus; constituent of 66S pre-ribosomal particles;
K14847 ribosome production factor 2
Length=344
Score = 67.0 bits (162), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 51/217 (23%), Positives = 82/217 (37%), Gaps = 53/217 (24%)
Query 1 FENAEPIEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVA-HYTPAAAF 59
FE+ P+E+ + KN C L +SSKKR + R + +I DM E VA ++ + F
Sbjct 69 FEDMSPLEFFSEKNDCSLMVLMTSSKKRKNNMTFIRTFGYKIYDMIELMVADNFKLLSDF 128
Query 60 AAVQAPAAGSAPLLLLQGGLWDVSDEMRNARNIFGDLFRAAEVPPDGPTQKLYLGGVDRL 119
+ G P+ QG +D + +++F D FR
Sbjct 129 KKLTF-TVGLKPMFTFQGAAFDTHPVYKQIKSLFLDFFRG-------------------- 167
Query 120 IAVSAVQATPGSAATAATTAAAAKAVIGGSEDCETEAPASGATTDGTDAPAADSGYSGDV 179
+T A + VI + G DG P
Sbjct 168 ------------ESTDLQDVAGLQHVISMT--------IQGDFQDGEPLPN--------- 198
Query 180 LICVRHYRIALVKTADAGGVGGPAVKLVEVGPQIDLR 216
+ R Y++ K +D GG P ++LVE+GP++D +
Sbjct 199 -VLFRVYKLKSYK-SDQGGKRLPRIELVEIGPRLDFK 233
> pfa:PF07_0122 nucleolus BRIX protein, putative; K14820 ribosome
biogenesis protein BRX1
Length=416
Score = 34.7 bits (78), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 26/52 (50%), Gaps = 5/52 (9%)
Query 168 APAADSGYSG---DVLICVRHYRIALVKTADAGGVGGPAVKLVEVGPQIDLR 216
P D Y+ + LI RHY+I V AD+ V +LVE+GPQ L
Sbjct 302 KPFYDHCYNFFYVNDLIYFRHYQILPVTLADSNNVNKQ--QLVEIGPQFTLH 351
> mmu:382051 Pdp2, 4833426J09Rik, Gm1705, KIAA1348, mKIAA1348;
pyruvate dehyrogenase phosphatase catalytic subunit 2 (EC:3.1.3.43);
K01102 pyruvate dehydrogenase phosphatase [EC:3.1.3.43]
Length=532
Score = 33.9 bits (76), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 36/134 (26%), Positives = 52/134 (38%), Gaps = 7/134 (5%)
Query 27 KRPARLVLGRVYDNQILDMQEFAVAHY-TP----AAAFAAVQAPAAGSAPLLLLQGGLWD 81
K R VL R +D + L++ +F HY TP A L+L GLWD
Sbjct 360 KELQRNVLARGFDTEALNIYQFTPPHYYTPPYLTAKPEVTYHRLRRQDKFLVLASDGLWD 419
Query 82 VSDEMRNARNIFGDLFRAAEVPPDGPTQKLYLGGVDRLIAVSAVQATPGSAATAATTAAA 141
+ R + G L + PD + LG + L+ +A+ AA T
Sbjct 420 MLGNEDVVRLVVGHLSKVGRHKPDLDQRPANLGLMQSLLLQR--KASGLHAADQNTATHL 477
Query 142 AKAVIGGSEDCETE 155
+ IG +E E E
Sbjct 478 IRHAIGSNEYGEME 491
> tgo:TGME49_094820 type I fatty acid synthase, putative (EC:6.2.1.3
1.1.1.1 1.2.1.31 1.1.1.9 2.3.1.94 1.1.1.36 2.3.1.161
2.3.1.86 2.3.1.39 2.3.1.16 2.8.2.20 1.6.5.5 1.1.1.14)
Length=10021
Score = 30.0 bits (66), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 34/127 (26%), Positives = 49/127 (38%), Gaps = 18/127 (14%)
Query 79 LWDVSDEMRNARNIFG---------DLFRAAEVPPDGPTQK------LYLGGVDRLIAVS 123
L ++D R + I G D A+ P+GP Q+ L +GGVD L
Sbjct 7244 LRHLADAQREGKRILGIVRGTAVNHDGRSASLTAPNGPAQQDVIRSALQIGGVDPLDVAL 7303
Query 124 AVQATPGSAATAATTAAAAKAVIGGSEDCETEAPASGATTDGTDAPAADSGYSG--DVLI 181
G+A A KAV G ++ GA SG +G VL+
Sbjct 7304 VESHGTGTALGDPIEMGAIKAVYGAGRSADSPL-VVGALKSYIGHLEGSSGIAGILKVLL 7362
Query 182 CVRHYRI 188
C+RH+ +
Sbjct 7363 CLRHHEV 7369
Lambda K H
0.316 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7009334380
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40