bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4471_orf1
Length=105
Score E
Sequences producing significant alignments: (Bits) Value
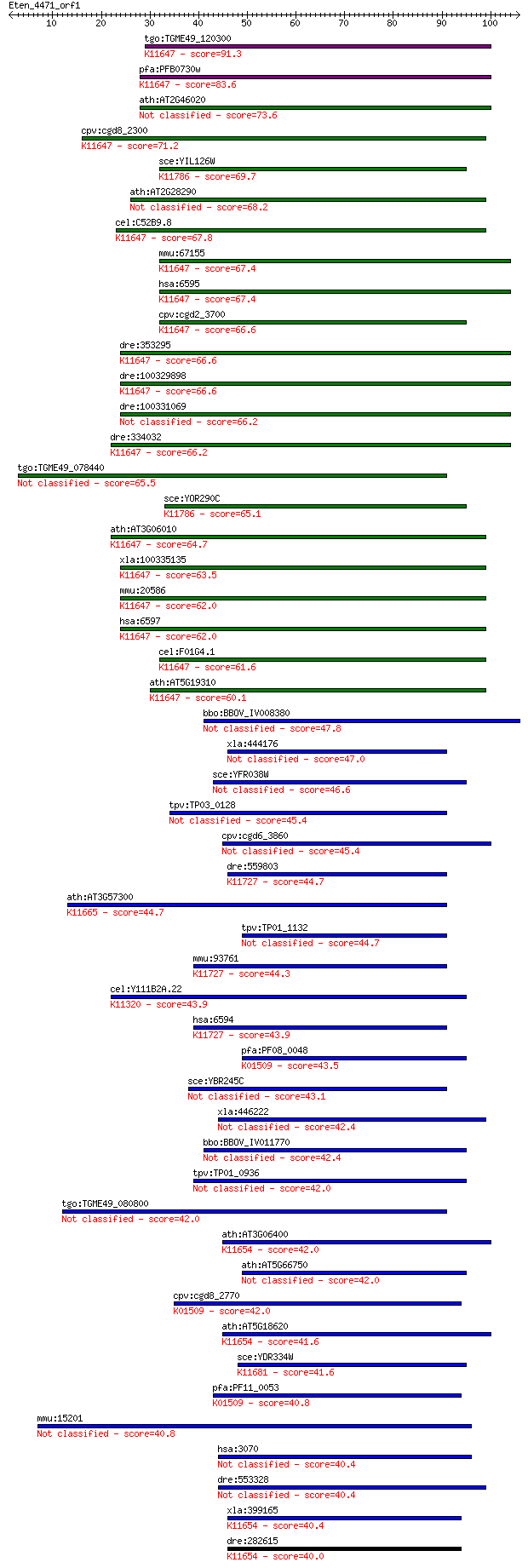
tgo:TGME49_120300 SNF2 family N-terminal domain-containing pro... 91.3 7e-19
pfa:PFB0730w DEAD/DEAH box helicase, putative; K11647 SWI/SNF-... 83.6 2e-16
ath:AT2G46020 BRM; transcription regulatory protein SNF2, puta... 73.6 2e-13
cpv:cgd8_2300 brahma like protein with a HSA domain, SNF2 like... 71.2 7e-13
sce:YIL126W STH1, NPS1; ATPase component of the RSC chromatin ... 69.7 2e-12
ath:AT2G28290 SYD; SYD (SPLAYED); ATPase/ chromatin binding 68.2 6e-12
cel:C52B9.8 hypothetical protein; K11647 SWI/SNF-related matri... 67.8 8e-12
mmu:67155 Smarca2, 2610209L14Rik, SNF2alpha, Snf2l2, brahma, b... 67.4 1e-11
hsa:6595 SMARCA2, BAF190, BRM, FLJ36757, MGC74511, SNF2, SNF2L... 67.4 1e-11
cpv:cgd2_3700 SWI/SNF related transcriptional regulator ATpase... 66.6 2e-11
dre:353295 smarca4, brg1, smarca2, wu:fb51a08; SWI/SNF related... 66.6 2e-11
dre:100329898 SWI/SNF-related matrix-associated actin-dependen... 66.6 2e-11
dre:100331069 SWI/SNF-related matrix-associated actin-dependen... 66.2 2e-11
dre:334032 smarca2, psa-4, wu:fa56c07, wu:fi27f11, zgc:66238; ... 66.2 2e-11
tgo:TGME49_078440 transcription regulatory protein SNF2, putat... 65.5 4e-11
sce:YOR290C SNF2, GAM1, HAF1, SWI2, TYE3; Catalytic subunit of... 65.1 6e-11
ath:AT3G06010 ATCHR12; ATP binding / DNA binding / helicase/ n... 64.7 7e-11
xla:100335135 smarca4, baf190, brg1, rtps2, snf2, snf2-beta, s... 63.5 1e-10
mmu:20586 Smarca4, Brg1, HP1-BP72, SNF2beta, SW1/SNF; SWI/SNF ... 62.0 4e-10
hsa:6597 SMARCA4, BAF190, BRG1, FLJ39786, RTPS2, SNF2, SNF2-BE... 62.0 4e-10
cel:F01G4.1 psa-4; Phasmid Socket Absent family member (psa-4)... 61.6 6e-10
ath:AT5G19310 homeotic gene regulator, putative; K11647 SWI/SN... 60.1 2e-09
bbo:BBOV_IV008380 23.m05834; SNF2 helicase (EC:3.6.1.-) 47.8 8e-06
xla:444176 smarca1, MGC80667, brg1, snf2l; SWI/SNF related, ma... 47.0 1e-05
sce:YFR038W IRC5; Irc5p (EC:3.6.1.-) 46.6 2e-05
tpv:TP03_0128 global transcription activator 45.4 4e-05
cpv:cgd6_3860 SNF2 helicase 45.4 4e-05
dre:559803 novel protein similar to SWI/SNF related, matrix as... 44.7 7e-05
ath:AT3G57300 INO80; INO80 (INO80 ORTHOLOG); ATP binding / DNA... 44.7 8e-05
tpv:TP01_1132 ATP-dependent helicase 44.7 8e-05
mmu:93761 Smarca1, 5730494M04Rik, Snf2l; SWI/SNF related, matr... 44.3 9e-05
cel:Y111B2A.22 ssl-1; yeast Swi2/Snf2-Like family member (ssl-... 43.9 1e-04
hsa:6594 SMARCA1, DKFZp686D1623, FLJ41547, ISWI, NURF140, SNF2... 43.9 1e-04
pfa:PF08_0048 ATP-dependent helicase, putative; K01509 adenosi... 43.5 2e-04
sce:YBR245C ISW1, SGN2; Isw1p (EC:3.6.1.-) 43.1 2e-04
xla:446222 hells, lsh, nbla10143, pasg, smarca6; helicase, lym... 42.4 3e-04
bbo:BBOV_IV011770 23.m06400; snf2-related chromatin remodeling... 42.4 4e-04
tpv:TP01_0936 DNA-dependent ATPase 42.0 4e-04
tgo:TGME49_080800 SNF2 family N-terminal domain-containing pro... 42.0 5e-04
ath:AT3G06400 CHR11; CHR11 (CHROMATIN-REMODELING PROTEIN 11); ... 42.0 5e-04
ath:AT5G66750 CHR1; CHR1 (CHROMATIN REMODELING 1); ATPase/ hel... 42.0 5e-04
cpv:cgd8_2770 SNF2L ortholog with a SWI/SNF2 like ATpase and a... 42.0 5e-04
ath:AT5G18620 CHR17; CHR17 (CHROMATIN REMODELING FACTOR17); AT... 41.6 6e-04
sce:YDR334W SWR1; Swi2/Snf2-related ATPase that is the structu... 41.6 6e-04
pfa:PF11_0053 PfSNF2L; K01509 adenosinetriphosphatase [EC:3.6.... 40.8 0.001
mmu:15201 Hells, AI323785, E130115I21Rik, LSH, Lysh, PASG, YFK... 40.8 0.001
hsa:3070 HELLS, FLJ10339, LSH, PASG, SMARCA6; helicase, lympho... 40.4 0.001
dre:553328 hells, cb65, im:6911667, pasg, sb:cb65, sb:cb749; h... 40.4 0.001
xla:399165 smarca5, iswi; SWI/SNF related, matrix associated, ... 40.4 0.001
dre:282615 smarca5, chunp6878, fb26d12, fb49g04, im:7146484, w... 40.0 0.002
> tgo:TGME49_120300 SNF2 family N-terminal domain-containing protein
(EC:2.7.11.1 3.2.1.3); K11647 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily
A member 2/4 [EC:3.6.4.-]
Length=1606
Score = 91.3 bits (225), Expect = 7e-19, Method: Composition-based stats.
Identities = 39/71 (54%), Positives = 53/71 (74%), Gaps = 0/71 (0%)
Query 29 RDRYYTLSHAVREEVRQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLM 88
+D+YY +SH V+EEV+QP +L GG LMPYQMAGL WMLSLYNN+LHGIL + L ++
Sbjct 665 QDQYYAMSHQVQEEVQQPSTLTGGDLMPYQMAGLSWMLSLYNNDLHGILADEMGLGKTIQ 724
Query 89 SVECISSSIEF 99
++ ++ EF
Sbjct 725 TIALLAYLKEF 735
> pfa:PFB0730w DEAD/DEAH box helicase, putative; K11647 SWI/SNF-related
matrix-associated actin-dependent regulator of chromatin
subfamily A member 2/4 [EC:3.6.4.-]
Length=1997
Score = 83.6 bits (205), Expect = 2e-16, Method: Composition-based stats.
Identities = 34/72 (47%), Positives = 51/72 (70%), Gaps = 0/72 (0%)
Query 28 SRDRYYTLSHAVREEVRQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSL 87
+R+ YY +SH V+E+V+QP L+GG LM YQ+ GL+W++SLYNNNLHGIL + L ++
Sbjct 858 ARENYYNISHVVKEKVKQPSILIGGELMKYQLEGLEWLVSLYNNNLHGILADEMGLGKTI 917
Query 88 MSVECISSSIEF 99
++ + EF
Sbjct 918 QTISLFAYLKEF 929
> ath:AT2G46020 BRM; transcription regulatory protein SNF2, putative
Length=2192
Score = 73.6 bits (179), Expect = 2e-13, Method: Composition-based stats.
Identities = 38/73 (52%), Positives = 49/73 (67%), Gaps = 1/73 (1%)
Query 28 SRDRYYTLSHAVREEV-RQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPS 86
S ++YYTL+HAV E V RQP L GTL YQ+ GLQWMLSLYNN L+GIL + L +
Sbjct 954 SVNKYYTLAHAVNEVVVRQPSMLQAGTLRDYQLVGLQWMLSLYNNKLNGILADEMGLGKT 1013
Query 87 LMSVECISSSIEF 99
+ + I+ +EF
Sbjct 1014 VQVMALIAYLMEF 1026
> cpv:cgd8_2300 brahma like protein with a HSA domain, SNF2 like
helicase and a bromo domain ; K11647 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily
A member 2/4 [EC:3.6.4.-]
Length=1673
Score = 71.2 bits (173), Expect = 7e-13, Method: Composition-based stats.
Identities = 34/91 (37%), Positives = 56/91 (61%), Gaps = 8/91 (8%)
Query 16 TEGPCGSNKG-------LGSRDRYYTLSHAVREEV-RQPESLVGGTLMPYQMAGLQWMLS 67
+E C NK + +++RY+ ++H ++E + +QPE L GG L YQM GL+W++S
Sbjct 699 SETCCSKNKKKKRSAPLIRAKERYFQVTHMIQEHITKQPECLKGGQLREYQMKGLEWLVS 758
Query 68 LYNNNLHGILGAFLRLAPSLMSVECISSSIE 98
LYNNNL+GIL + L ++ +V ++ E
Sbjct 759 LYNNNLNGILADAMGLGKTVQTVSVLAHIYE 789
> sce:YIL126W STH1, NPS1; ATPase component of the RSC chromatin
remodeling complex; required for expression of early meiotic
genes; essential helicase-related protein homologous to Snf2p
(EC:3.6.1.-); K11786 ATP-dependent helicase STH1/SNF2 [EC:3.6.4.-]
Length=1359
Score = 69.7 bits (169), Expect = 2e-12, Method: Composition-based stats.
Identities = 30/64 (46%), Positives = 47/64 (73%), Gaps = 1/64 (1%)
Query 32 YYTLSHAVREEV-RQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSV 90
YY ++H ++E++ +QP LVGGTL YQ+ GL+WM+SLYNN+L+GIL + L ++ S+
Sbjct 447 YYEVAHRIKEKIDKQPSILVGGTLKEYQLRGLEWMVSLYNNHLNGILADEMGLGKTIQSI 506
Query 91 ECIS 94
I+
Sbjct 507 SLIT 510
> ath:AT2G28290 SYD; SYD (SPLAYED); ATPase/ chromatin binding
Length=3543
Score = 68.2 bits (165), Expect = 6e-12, Method: Composition-based stats.
Identities = 32/74 (43%), Positives = 50/74 (67%), Gaps = 1/74 (1%)
Query 26 LGSRDRYYTLSHAVREEV-RQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLA 84
L S ++YY ++H+++E + QP SLVGG L YQM GL+W++SLYNN+L+GIL + L
Sbjct 725 LESNEKYYLMAHSIKENINEQPSSLVGGKLREYQMNGLRWLVSLYNNHLNGILADEMGLG 784
Query 85 PSLMSVECISSSIE 98
++ + I +E
Sbjct 785 KTVQVISLICYLME 798
> cel:C52B9.8 hypothetical protein; K11647 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily
A member 2/4 [EC:3.6.4.-]
Length=1336
Score = 67.8 bits (164), Expect = 8e-12, Method: Composition-based stats.
Identities = 32/81 (39%), Positives = 52/81 (64%), Gaps = 5/81 (6%)
Query 23 NKGLGSRDRYYTLSHAVREEVRQPESLVGG-----TLMPYQMAGLQWMLSLYNNNLHGIL 77
NK + + YYT +H VREE+++ ++GG L PYQ+ GL+WM+SL+NNNL+GIL
Sbjct 330 NKTKMNIEDYYTTAHGVREEIKEQHFMMGGGNPSLKLKPYQIKGLEWMVSLFNNNLNGIL 389
Query 78 GAFLRLAPSLMSVECISSSIE 98
+ L ++ ++ I+ +E
Sbjct 390 ADEMGLGKTIQTIAFITYLME 410
> mmu:67155 Smarca2, 2610209L14Rik, SNF2alpha, Snf2l2, brahma,
brm; SWI/SNF related, matrix associated, actin dependent regulator
of chromatin, subfamily a, member 2; K11647 SWI/SNF-related
matrix-associated actin-dependent regulator of chromatin
subfamily A member 2/4 [EC:3.6.4.-]
Length=1583
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 32/73 (43%), Positives = 49/73 (67%), Gaps = 1/73 (1%)
Query 32 YYTLSHAVREEV-RQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSV 90
YYT++HA+ E V +Q L+ GTL YQ+ GL+WM+SLYNNNL+GIL + L ++ ++
Sbjct 712 YYTVAHAISERVEKQSALLINGTLKHYQLQGLEWMVSLYNNNLNGILADEMGLGKTIQTI 771
Query 91 ECISSSIEFTPLD 103
I+ +E L+
Sbjct 772 ALITYLMEHKRLN 784
> hsa:6595 SMARCA2, BAF190, BRM, FLJ36757, MGC74511, SNF2, SNF2L2,
SNF2LA, SWI2, Sth1p, hBRM, hSNF2a; SWI/SNF related, matrix
associated, actin dependent regulator of chromatin, subfamily
a, member 2; K11647 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily A member 2/4
[EC:3.6.4.-]
Length=1590
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 32/73 (43%), Positives = 49/73 (67%), Gaps = 1/73 (1%)
Query 32 YYTLSHAVREEV-RQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSV 90
YYT++HA+ E V +Q L+ GTL YQ+ GL+WM+SLYNNNL+GIL + L ++ ++
Sbjct 701 YYTVAHAISERVEKQSALLINGTLKHYQLQGLEWMVSLYNNNLNGILADEMGLGKTIQTI 760
Query 91 ECISSSIEFTPLD 103
I+ +E L+
Sbjct 761 ALITYLMEHKRLN 773
> cpv:cgd2_3700 SWI/SNF related transcriptional regulator ATpase
; K11647 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin subfamily A member 2/4 [EC:3.6.4.-]
Length=1552
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 29/65 (44%), Positives = 47/65 (72%), Gaps = 2/65 (3%)
Query 32 YYTLSHAVREEVR-QPESLV-GGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMS 89
YYT++H+V E + +P L+ GG+L+PYQ+ G++WMLSLYNN LHGIL + L ++ +
Sbjct 554 YYTMAHSVSESISDKPMKLLKGGSLLPYQIIGVEWMLSLYNNKLHGILADEMGLGKTVQT 613
Query 90 VECIS 94
+ ++
Sbjct 614 IALLT 618
> dre:353295 smarca4, brg1, smarca2, wu:fb51a08; SWI/SNF related,
matrix associated, actin dependent regulator of chromatin,
subfamily a, member 4; K11647 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily A member
2/4 [EC:3.6.4.-]
Length=1627
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 35/81 (43%), Positives = 53/81 (65%), Gaps = 4/81 (4%)
Query 24 KGLGSRDRYYTLSHAVREEV-RQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLR 82
+GL S YY ++HAV E+V +Q LV G L YQ+ GL+W++SLYNNNL+GIL +
Sbjct 738 RGLQS---YYAVAHAVTEKVEKQSSLLVNGQLKQYQIKGLEWLVSLYNNNLNGILADEMG 794
Query 83 LAPSLMSVECISSSIEFTPLD 103
L ++ ++ I+ +EF L+
Sbjct 795 LGKTIQTIALITYLMEFKRLN 815
> dre:100329898 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin a4-like; K11647 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily
A member 2/4 [EC:3.6.4.-]
Length=1627
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 35/81 (43%), Positives = 53/81 (65%), Gaps = 4/81 (4%)
Query 24 KGLGSRDRYYTLSHAVREEV-RQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLR 82
+GL S YY ++HAV E+V +Q LV G L YQ+ GL+W++SLYNNNL+GIL +
Sbjct 738 RGLQS---YYAVAHAVTEKVEKQSSLLVNGQLKQYQIKGLEWLVSLYNNNLNGILADEMG 794
Query 83 LAPSLMSVECISSSIEFTPLD 103
L ++ ++ I+ +EF L+
Sbjct 795 LGKTIQTIALITYLMEFKRLN 815
> dre:100331069 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin a4-like
Length=1234
Score = 66.2 bits (160), Expect = 2e-11, Method: Composition-based stats.
Identities = 35/81 (43%), Positives = 53/81 (65%), Gaps = 4/81 (4%)
Query 24 KGLGSRDRYYTLSHAVREEV-RQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLR 82
+GL S YY ++HAV E+V +Q LV G L YQ+ GL+W++SLYNNNL+GIL +
Sbjct 345 RGLQS---YYAVAHAVTEKVEKQSSLLVNGQLKQYQIKGLEWLVSLYNNNLNGILADEMG 401
Query 83 LAPSLMSVECISSSIEFTPLD 103
L ++ ++ I+ +EF L+
Sbjct 402 LGKTIQTIALITYLMEFKRLN 422
> dre:334032 smarca2, psa-4, wu:fa56c07, wu:fi27f11, zgc:66238;
SWI/SNF related, matrix associated, actin dependent regulator
of chromatin, subfamily a, member 2; K11647 SWI/SNF-related
matrix-associated actin-dependent regulator of chromatin
subfamily A member 2/4 [EC:3.6.4.-]
Length=1568
Score = 66.2 bits (160), Expect = 2e-11, Method: Composition-based stats.
Identities = 35/83 (42%), Positives = 51/83 (61%), Gaps = 1/83 (1%)
Query 22 SNKGLGSRDRYYTLSHAVREEV-RQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAF 80
S G S YY ++HAV E V +Q L+ GTL YQ+ GL+WM+SLYNNNL+GIL
Sbjct 693 SQAGQTSSQSYYGVAHAVIERVDKQSTFLINGTLKQYQIQGLEWMVSLYNNNLNGILADE 752
Query 81 LRLAPSLMSVECISSSIEFTPLD 103
+ L ++ ++ I+ +E L+
Sbjct 753 MGLGKTIQTIGLITYLMEHKRLN 775
> tgo:TGME49_078440 transcription regulatory protein SNF2, putative
(EC:2.7.11.1 3.4.21.97 3.2.1.3 3.1.3.33 3.4.24.61 3.4.24.35)
Length=2668
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 38/91 (41%), Positives = 53/91 (58%), Gaps = 3/91 (3%)
Query 3 LRSGDG--GDSTTTITEGPCGSNKGLGSRDRYYTLSHAVREEVRQ-PESLVGGTLMPYQM 59
+ GDG GD+T + + L S++RYY L+HA R V + P+ L GG+L YQM
Sbjct 1165 MEQGDGKVGDATDEEEKNKASLSSFLLSKERYYRLTHAKRVHVTELPKCLKGGSLRSYQM 1224
Query 60 AGLQWMLSLYNNNLHGILGAFLRLAPSLMSV 90
GL WM SLY N L+GIL + L ++ +V
Sbjct 1225 EGLNWMASLYTNGLNGILADSMGLGKTVQTV 1255
> sce:YOR290C SNF2, GAM1, HAF1, SWI2, TYE3; Catalytic subunit
of the SWI/SNF chromatin remodeling complex involved in transcriptional
regulation; contains DNA-stimulated ATPase activity;
functions interdependently in transcriptional activation
with Snf5p and Snf6p (EC:3.6.1.-); K11786 ATP-dependent helicase
STH1/SNF2 [EC:3.6.4.-]
Length=1703
Score = 65.1 bits (157), Expect = 6e-11, Method: Composition-based stats.
Identities = 27/63 (42%), Positives = 46/63 (73%), Gaps = 1/63 (1%)
Query 33 YTLSHAVREEVR-QPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVE 91
Y ++H ++E+++ QP LVGGTL YQ+ GLQWM+SL+NN+L+GIL + L ++ ++
Sbjct 745 YNVAHRIKEDIKKQPSILVGGTLKDYQIKGLQWMVSLFNNHLNGILADEMGLGKTIQTIS 804
Query 92 CIS 94
++
Sbjct 805 LLT 807
> ath:AT3G06010 ATCHR12; ATP binding / DNA binding / helicase/
nucleic acid binding; K11647 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily A member
2/4 [EC:3.6.4.-]
Length=1132
Score = 64.7 bits (156), Expect = 7e-11, Method: Composition-based stats.
Identities = 32/78 (41%), Positives = 50/78 (64%), Gaps = 1/78 (1%)
Query 22 SNKGLGSRDRYYTLSHAVREEV-RQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAF 80
SN L + +Y + H+++E+V QP L GG L YQ+ GLQWM+SL+NNNL+GIL
Sbjct 400 SNDLLEGQRQYNSAIHSIQEKVTEQPSLLEGGELRSYQLEGLQWMVSLFNNNLNGILADE 459
Query 81 LRLAPSLMSVECISSSIE 98
+ L ++ ++ I+ +E
Sbjct 460 MGLGKTIQTISLIAYLLE 477
> xla:100335135 smarca4, baf190, brg1, rtps2, snf2, snf2-beta,
snf2b, snf2l4, snf2lb, swi2; SWI/SNF related, matrix associated,
actin dependent regulator of chromatin, subfamily a, member
4; K11647 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin subfamily A member 2/4 [EC:3.6.4.-]
Length=1600
Score = 63.5 bits (153), Expect = 1e-10, Method: Composition-based stats.
Identities = 33/76 (43%), Positives = 50/76 (65%), Gaps = 4/76 (5%)
Query 24 KGLGSRDRYYTLSHAVREEV-RQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLR 82
+GL S YY ++HAV E+V +Q LV G L YQ+ GL+W++SLYNNNL+GIL +
Sbjct 711 RGLQS---YYAVAHAVSEKVEKQSSLLVNGILKQYQIKGLEWLVSLYNNNLNGILADEMG 767
Query 83 LAPSLMSVECISSSIE 98
L ++ ++ I+ +E
Sbjct 768 LGKTIQTIALITYLME 783
> mmu:20586 Smarca4, Brg1, HP1-BP72, SNF2beta, SW1/SNF; SWI/SNF
related, matrix associated, actin dependent regulator of chromatin,
subfamily a, member 4; K11647 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily
A member 2/4 [EC:3.6.4.-]
Length=1617
Score = 62.0 bits (149), Expect = 4e-10, Method: Composition-based stats.
Identities = 32/76 (42%), Positives = 49/76 (64%), Gaps = 4/76 (5%)
Query 24 KGLGSRDRYYTLSHAVREEV-RQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLR 82
+GL S YY ++HAV E V +Q +V G L YQ+ GL+W++SLYNNNL+GIL +
Sbjct 726 RGLQS---YYAVAHAVTERVDKQSALMVNGVLKQYQIKGLEWLVSLYNNNLNGILADEMG 782
Query 83 LAPSLMSVECISSSIE 98
L ++ ++ I+ +E
Sbjct 783 LGKTIQTIALITYLME 798
> hsa:6597 SMARCA4, BAF190, BRG1, FLJ39786, RTPS2, SNF2, SNF2-BETA,
SNF2L4, SNF2LB, SWI2, hSNF2b; SWI/SNF related, matrix
associated, actin dependent regulator of chromatin, subfamily
a, member 4; K11647 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin subfamily A member 2/4 [EC:3.6.4.-]
Length=1647
Score = 62.0 bits (149), Expect = 4e-10, Method: Composition-based stats.
Identities = 32/76 (42%), Positives = 49/76 (64%), Gaps = 4/76 (5%)
Query 24 KGLGSRDRYYTLSHAVREEV-RQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLR 82
+GL S YY ++HAV E V +Q +V G L YQ+ GL+W++SLYNNNL+GIL +
Sbjct 726 RGLQS---YYAVAHAVTERVDKQSALMVNGVLKQYQIKGLEWLVSLYNNNLNGILADEMG 782
Query 83 LAPSLMSVECISSSIE 98
L ++ ++ I+ +E
Sbjct 783 LGKTIQTIALITYLME 798
> cel:F01G4.1 psa-4; Phasmid Socket Absent family member (psa-4);
K11647 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin subfamily A member 2/4 [EC:3.6.4.-]
Length=1474
Score = 61.6 bits (148), Expect = 6e-10, Method: Composition-based stats.
Identities = 27/72 (37%), Positives = 46/72 (63%), Gaps = 5/72 (6%)
Query 32 YYTLSHAVREEVRQPESLVGG-----TLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPS 86
YY +H ++E+V + + +GG L PYQ+ GL+WM+SLYNNNL+GIL + L +
Sbjct 506 YYATAHKIKEKVVKQHTTMGGGDPNLLLKPYQIKGLEWMVSLYNNNLNGILADEMGLGKT 565
Query 87 LMSVECISSSIE 98
+ ++ ++ +E
Sbjct 566 IQTISLVTYLME 577
> ath:AT5G19310 homeotic gene regulator, putative; K11647 SWI/SNF-related
matrix-associated actin-dependent regulator of chromatin
subfamily A member 2/4 [EC:3.6.4.-]
Length=1064
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 29/71 (40%), Positives = 48/71 (67%), Gaps = 2/71 (2%)
Query 30 DRYYTLS-HAVREEV-RQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSL 87
+R + L+ H+++E+V +QP L GG L YQ+ GLQWM+SLYNN+ +GIL + L ++
Sbjct 360 ERQFNLAIHSIQEKVTKQPSLLQGGELRSYQLEGLQWMVSLYNNDYNGILADEMGLGKTI 419
Query 88 MSVECISSSIE 98
++ I+ +E
Sbjct 420 QTIALIAYLLE 430
> bbo:BBOV_IV008380 23.m05834; SNF2 helicase (EC:3.6.1.-)
Length=894
Score = 47.8 bits (112), Expect = 8e-06, Method: Composition-based stats.
Identities = 22/65 (33%), Positives = 40/65 (61%), Gaps = 1/65 (1%)
Query 41 EEVRQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECISSSIEFT 100
E +QP++LVG T PYQ+ GL+W++ LY+ N++GIL + L + ++ ++ E
Sbjct 74 EVTQQPKNLVG-TAKPYQLEGLRWLVGLYDRNMNGILADEMGLGKTFQTISLLAYLKESR 132
Query 101 PLDSV 105
+D +
Sbjct 133 GIDGL 137
> xla:444176 smarca1, MGC80667, brg1, snf2l; SWI/SNF related,
matrix associated, actin dependent regulator of chromatin, subfamily
a, member 1
Length=403
Score = 47.0 bits (110), Expect = 1e-05, Method: Composition-based stats.
Identities = 20/45 (44%), Positives = 29/45 (64%), Gaps = 0/45 (0%)
Query 46 PESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSV 90
P + GGTL YQ+ GL WM+SLY N ++GIL + L +L ++
Sbjct 152 PSYIKGGTLRDYQVRGLNWMISLYENGINGILADEMGLGKTLQTI 196
> sce:YFR038W IRC5; Irc5p (EC:3.6.1.-)
Length=853
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 32/52 (61%), Gaps = 0/52 (0%)
Query 43 VRQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECIS 94
++QP L L PYQ+ GL W+++LY N L+GIL + L ++ S+ ++
Sbjct 211 IKQPRLLKNCILKPYQLEGLNWLITLYENGLNGILADEMGLGKTVQSIALLA 262
> tpv:TP03_0128 global transcription activator
Length=998
Score = 45.4 bits (106), Expect = 4e-05, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 37/59 (62%), Gaps = 3/59 (5%)
Query 34 TLSHAVREEVRQ--PESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSV 90
T+ + +RE + P +L+G L YQ+ GL W++SLYNN L+GIL + L ++ ++
Sbjct 422 TVEYIIRENIFNNIPNALIG-KLRNYQLYGLDWLVSLYNNKLNGILADEMGLGKTIQTI 479
> cpv:cgd6_3860 SNF2 helicase
Length=1102
Score = 45.4 bits (106), Expect = 4e-05, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 31/55 (56%), Gaps = 0/55 (0%)
Query 45 QPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECISSSIEF 99
QP + G L PYQ+ GL W+++LY L+GIL + L + S+ ++ E+
Sbjct 176 QPACIQNGVLKPYQLEGLNWLINLYEGGLNGILADEMGLGKTFQSISLLAYLREY 230
> dre:559803 novel protein similar to SWI/SNF related, matrix
associated, actin dependent regulator of chromatin, subfamily
a, member 5 (smarca5); K11727 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily A member
1 [EC:3.6.4.-]
Length=1036
Score = 44.7 bits (104), Expect = 7e-05, Method: Composition-based stats.
Identities = 19/45 (42%), Positives = 28/45 (62%), Gaps = 0/45 (0%)
Query 46 PESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSV 90
P + GTL YQ+ GL WM+SLY N ++GIL + L +L ++
Sbjct 135 PSYIKNGTLRDYQIRGLNWMISLYENGINGILADEMGLGKTLQTI 179
> ath:AT3G57300 INO80; INO80 (INO80 ORTHOLOG); ATP binding / DNA
binding / helicase/ nucleic acid binding; K11665 DNA helicase
INO80 [EC:3.6.4.12]
Length=1507
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 28/80 (35%), Positives = 42/80 (52%), Gaps = 3/80 (3%)
Query 13 TTITEGPCG--SNKGLGSRDRYYTLSHAVREEVRQPESLVGGTLMPYQMAGLQWMLSLYN 70
T+ EGP S G + D + + V V+ PE L GTL YQM GLQW+++ Y
Sbjct 544 TSEMEGPLNDISVSGSSNIDLHNPSTMPVTSTVQTPE-LFKGTLKEYQMKGLQWLVNCYE 602
Query 71 NNLHGILGAFLRLAPSLMSV 90
L+GIL + L ++ ++
Sbjct 603 QGLNGILADEMGLGKTIQAM 622
> tpv:TP01_1132 ATP-dependent helicase
Length=1632
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 18/42 (42%), Positives = 28/42 (66%), Gaps = 0/42 (0%)
Query 49 LVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSV 90
L+ G L PYQ GL+W++SLY N++GIL + L +L ++
Sbjct 693 LIKGVLRPYQKEGLRWLVSLYERNINGILADEMGLGKTLQTI 734
> mmu:93761 Smarca1, 5730494M04Rik, Snf2l; SWI/SNF related, matrix
associated, actin dependent regulator of chromatin, subfamily
a, member 1; K11727 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily A member 1
[EC:3.6.4.-]
Length=1046
Score = 44.3 bits (103), Expect = 9e-05, Method: Composition-based stats.
Identities = 22/52 (42%), Positives = 32/52 (61%), Gaps = 1/52 (1%)
Query 39 VREEVRQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSV 90
VR EV P + GG L YQ+ GL W++SLY N ++GIL + L +L ++
Sbjct 173 VRFEV-SPSYVKGGPLRDYQIRGLNWLISLYENGVNGILADEMGLGKTLQTI 223
> cel:Y111B2A.22 ssl-1; yeast Swi2/Snf2-Like family member (ssl-1);
K11320 E1A-binding protein p400 [EC:3.6.4.-]
Length=2395
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 24/74 (32%), Positives = 41/74 (55%), Gaps = 4/74 (5%)
Query 22 SNKGLGSRDRYYTLSHAVREEVRQPES-LVGGTLMPYQMAGLQWMLSLYNNNLHGILGAF 80
+ + L + + YTL +V+ P L+ G L YQM GL WM++LY NL+GIL
Sbjct 528 AEEALKFQPKGYTLETT---QVKTPVPFLIRGQLREYQMVGLDWMVTLYEKNLNGILADE 584
Query 81 LRLAPSLMSVECIS 94
+ L ++ ++ ++
Sbjct 585 MGLGKTIQTISLLA 598
> hsa:6594 SMARCA1, DKFZp686D1623, FLJ41547, ISWI, NURF140, SNF2L,
SNF2L1, SNF2LB, SNF2LT, SWI, SWI2; SWI/SNF related, matrix
associated, actin dependent regulator of chromatin, subfamily
a, member 1; K11727 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily A member 1
[EC:3.6.4.-]
Length=1054
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 21/52 (40%), Positives = 32/52 (61%), Gaps = 1/52 (1%)
Query 39 VREEVRQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSV 90
+R EV P + GG L YQ+ GL W++SLY N ++GIL + L +L ++
Sbjct 169 IRFEV-SPSYVKGGPLRDYQIRGLNWLISLYENGVNGILADEMGLGKTLQTI 219
> pfa:PF08_0048 ATP-dependent helicase, putative; K01509 adenosinetriphosphatase
[EC:3.6.1.3]
Length=2082
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 21/46 (45%), Positives = 29/46 (63%), Gaps = 3/46 (6%)
Query 49 LVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECIS 94
++ TL YQ AGL W+L LY NN++GIL + L +L +CIS
Sbjct 657 IIKATLRDYQHAGLHWLLYLYKNNINGILADEMGLGKTL---QCIS 699
> sce:YBR245C ISW1, SGN2; Isw1p (EC:3.6.1.-)
Length=1129
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 32/53 (60%), Gaps = 0/53 (0%)
Query 38 AVREEVRQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSV 90
++ + R+ + V G L PYQ+ G+ W++SL+ N + GIL + L +L ++
Sbjct 180 SIEFQFRESPAYVNGQLRPYQIQGVNWLVSLHKNKIAGILADEMGLGKTLQTI 232
> xla:446222 hells, lsh, nbla10143, pasg, smarca6; helicase, lymphoid-specific
Length=838
Score = 42.4 bits (98), Expect = 3e-04, Method: Composition-based stats.
Identities = 17/55 (30%), Positives = 32/55 (58%), Gaps = 0/55 (0%)
Query 44 RQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECISSSIE 98
+QP+ GG + YQ+ G++W+ L+ N ++GIL + L ++ + IS +E
Sbjct 211 QQPKYFTGGVMRWYQIEGMEWLRMLWENGINGILADEMGLGKTVQCIATISMMVE 265
> bbo:BBOV_IV011770 23.m06400; snf2-related chromatin remodeling
factor SRCAP
Length=1675
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 33/54 (61%), Gaps = 1/54 (1%)
Query 41 EEVRQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECIS 94
+EV+ P L+ L PYQ+ GL+W+ SLY N +GIL + L +L ++ ++
Sbjct 661 DEVQVP-CLIRAVLRPYQLDGLRWLASLYRNKSNGILADEMGLGKTLQTIALLA 713
> tpv:TP01_0936 DNA-dependent ATPase
Length=1253
Score = 42.0 bits (97), Expect = 4e-04, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 34/56 (60%), Gaps = 1/56 (1%)
Query 39 VREEVRQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECIS 94
+ E V QP+ LVG + PYQ+ GL+W++ LY L+GIL + L + ++ ++
Sbjct 161 IVEAVDQPKILVGQS-KPYQIEGLKWLVGLYVKGLNGILADEMGLGKTFQTISFLA 215
> tgo:TGME49_080800 SNF2 family N-terminal domain-containing protein
(EC:2.7.11.1 2.7.1.127)
Length=2894
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 45/87 (51%), Gaps = 10/87 (11%)
Query 12 TTTITEGPCGSNKGLGSRDRYYTLSHAVREEVRQPE--------SLVGGTLMPYQMAGLQ 63
++ +++GP G +G S ++ + S E QP +LV TL YQ G+Q
Sbjct 1173 SSALSQGPHGEGRGGQSSEKTKSASDT--EPSPQPRYLSSNPAPALVRATLRTYQSEGVQ 1230
Query 64 WMLSLYNNNLHGILGAFLRLAPSLMSV 90
W+ +L++ L+GIL + L +L ++
Sbjct 1231 WLFALHDKGLNGILADEMGLGKTLQTI 1257
> ath:AT3G06400 CHR11; CHR11 (CHROMATIN-REMODELING PROTEIN 11);
ATP binding / DNA binding / DNA-dependent ATPase/ helicase/
hydrolase, acting on acid anhydrides, in phosphorus-containing
anhydrides / nucleic acid binding / nucleosome binding;
K11654 SWI/SNF-related matrix-associated actin-dependent regulator
of chromatin subfamily A member 5 [EC:3.6.4.-]
Length=1055
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 33/55 (60%), Gaps = 1/55 (1%)
Query 45 QPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECISSSIEF 99
QP S + G + YQ+AGL W++ LY N ++GIL + L +L ++ ++ E+
Sbjct 181 QP-SCIQGKMRDYQLAGLNWLIRLYENGINGILADEMGLGKTLQTISLLAYLHEY 234
> ath:AT5G66750 CHR1; CHR1 (CHROMATIN REMODELING 1); ATPase/ helicase
Length=764
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 30/46 (65%), Gaps = 0/46 (0%)
Query 49 LVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECIS 94
L GG L YQ+ G++W++SL+ N L+GIL + L ++ ++ +S
Sbjct 197 LTGGQLKSYQLKGVKWLISLWQNGLNGILADQMGLGKTIQTIGFLS 242
> cpv:cgd8_2770 SNF2L ortholog with a SWI/SNF2 like ATpase and
a Myb domain ; K01509 adenosinetriphosphatase [EC:3.6.1.3]
Length=1308
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 17/59 (28%), Positives = 35/59 (59%), Gaps = 1/59 (1%)
Query 35 LSHAVREEVRQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECI 93
+++++ + QP+ + G + YQ+ GL WM LY +N++GIL + L +L ++ +
Sbjct 146 INYSIEKVAEQPDCITG-KMKFYQLEGLNWMFQLYKHNINGILADEMGLGKTLQTISIL 203
> ath:AT5G18620 CHR17; CHR17 (CHROMATIN REMODELING FACTOR17);
ATP binding / DNA binding / DNA-dependent ATPase/ helicase/
hydrolase, acting on acid anhydrides, in phosphorus-containing
anhydrides / nucleic acid binding / nucleosome binding; K11654
SWI/SNF-related matrix-associated actin-dependent regulator
of chromatin subfamily A member 5 [EC:3.6.4.-]
Length=1069
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 33/55 (60%), Gaps = 1/55 (1%)
Query 45 QPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECISSSIEF 99
QP + + G L YQ+AGL W++ LY N ++GIL + L +L ++ ++ E+
Sbjct 186 QP-ACIQGKLRDYQLAGLNWLIRLYENGINGILADEMGLGKTLQTISLLAYLHEY 239
> sce:YDR334W SWR1; Swi2/Snf2-related ATPase that is the structural
component of the SWR1 complex, which exchanges histone
variant H2AZ (Htz1p) for chromatin-bound histone H2A (EC:3.6.1.-);
K11681 helicase SWR1 [EC:3.6.4.12]
Length=1514
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 18/47 (38%), Positives = 29/47 (61%), Gaps = 0/47 (0%)
Query 48 SLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECIS 94
SL+ G L YQ GL W+ SLYNN+ +GIL + L ++ ++ ++
Sbjct 690 SLLRGNLRTYQKQGLNWLASLYNNHTNGILADEMGLGKTIQTISLLA 736
> pfa:PF11_0053 PfSNF2L; K01509 adenosinetriphosphatase [EC:3.6.1.3]
Length=1426
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 31/51 (60%), Gaps = 1/51 (1%)
Query 43 VRQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECI 93
++QP + + GT+ PYQ+ GL W+ LY ++GIL + L +L ++ +
Sbjct 312 LKQPMN-INGTMKPYQLEGLNWLYQLYRFKINGILADEMGLGKTLQTISLL 361
> mmu:15201 Hells, AI323785, E130115I21Rik, LSH, Lysh, PASG, YFK8;
helicase, lymphoid specific
Length=821
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 30/108 (27%), Positives = 49/108 (45%), Gaps = 25/108 (23%)
Query 7 DGGDSTTTITEGPCGSNKGLGS--RDRYYTLSHAVREEVR-----------------QPE 47
D STT++ NK S +DR LS VR+ + QP+
Sbjct 143 DESSSTTSLCVEDIQKNKDSNSMIKDR---LSQTVRQNSKFFFDPVRKCNGQPVPFQQPK 199
Query 48 SLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECISS 95
GG + YQ+ G++W+ L+ N ++GIL + L +V+CI++
Sbjct 200 HFTGGVMRWYQVEGMEWLRMLWENGINGILADEMGLGK---TVQCIAT 244
> hsa:3070 HELLS, FLJ10339, LSH, PASG, SMARCA6; helicase, lymphoid-specific
Length=838
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 32/52 (61%), Gaps = 3/52 (5%)
Query 44 RQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECISS 95
+QP+ GG + YQ+ G++W+ L+ N ++GIL + L +V+CI++
Sbjct 213 QQPKHFTGGVMRWYQVEGMEWLRMLWENGINGILADEMGLGK---TVQCIAT 261
> dre:553328 hells, cb65, im:6911667, pasg, sb:cb65, sb:cb749;
helicase, lymphoid-specific
Length=853
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 16/55 (29%), Positives = 32/55 (58%), Gaps = 0/55 (0%)
Query 44 RQPESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECISSSIE 98
+QP+ GG + YQ+ G++W+ L+ N ++GIL + L ++ + I+ +E
Sbjct 223 QQPQLFTGGVMRWYQVEGIEWLRMLWENGINGILADEMGLGKTIQCIAHIAMMVE 277
> xla:399165 smarca5, iswi; SWI/SNF related, matrix associated,
actin dependent regulator of chromatin, subfamily a, member
5; K11654 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin subfamily A member 5 [EC:3.6.4.-]
Length=1046
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 28/48 (58%), Gaps = 0/48 (0%)
Query 46 PESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECI 93
P + G L YQ+ GL W++SLY N ++GIL + L +L ++ +
Sbjct 165 PAYVKSGKLRDYQVRGLNWLISLYENGINGILADEMGLGKTLQTISLL 212
> dre:282615 smarca5, chunp6878, fb26d12, fb49g04, im:7146484,
wu:fb26d12, wu:fb49g04, zgc:158434; SWI/SNF related, matrix
associated, actin dependent regulator of chromatin, subfamily
a, member 5; K11654 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin subfamily A member 5 [EC:3.6.4.-]
Length=1028
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 28/48 (58%), Gaps = 0/48 (0%)
Query 46 PESLVGGTLMPYQMAGLQWMLSLYNNNLHGILGAFLRLAPSLMSVECI 93
P + G L YQ+ GL W++SLY N ++GIL + L +L ++ +
Sbjct 148 PSYVKTGKLRDYQVRGLNWLISLYENGINGILADEMGLGKTLQTISLL 195
Lambda K H
0.317 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2017521068
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40