bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4473_orf1
Length=162
Score E
Sequences producing significant alignments: (Bits) Value
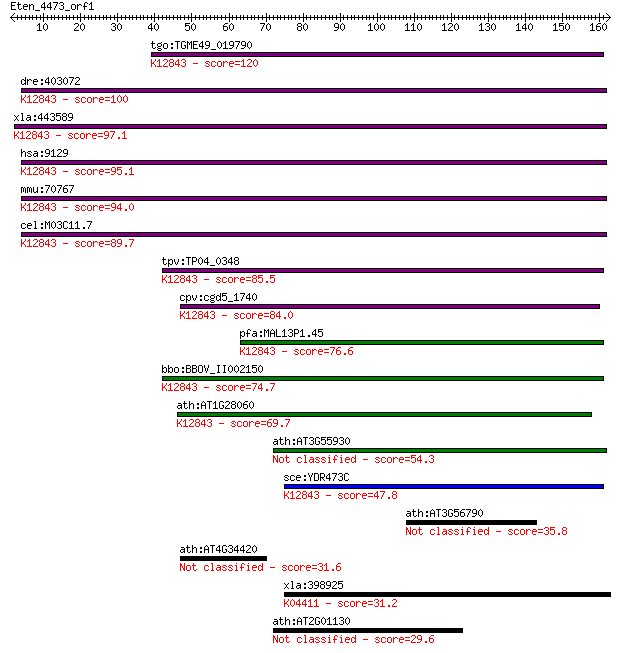
tgo:TGME49_019790 U4/U6 small nuclear ribonucleoprotein, putat... 120 2e-27
dre:403072 prpf3, HPRP3, HPRP3P; PRP3 pre-mRNA processing fact... 100 2e-21
xla:443589 prpf3, MGC130724; PRP3 pre-mRNA processing factor 3... 97.1 2e-20
hsa:9129 PRPF3, HPRP3, HPRP3P, PRP3, Prp3p, RP18; PRP3 pre-mRN... 95.1 8e-20
mmu:70767 Prpf3, 3632413F13Rik; PRP3 pre-mRNA processing facto... 94.0 2e-19
cel:M03C11.7 hypothetical protein; K12843 U4/U6 small nuclear ... 89.7 4e-18
tpv:TP04_0348 hypothetical protein; K12843 U4/U6 small nuclear... 85.5 8e-17
cpv:cgd5_1740 conserved protein, possible U4/U6 associated RNA... 84.0 2e-16
pfa:MAL13P1.45 U4/U6 small nuclear ribonucleoprotein, putative... 76.6 3e-14
bbo:BBOV_II002150 18.m06172; hypothetical protein; K12843 U4/U... 74.7 1e-13
ath:AT1G28060 small nuclear ribonucleoprotein family protein /... 69.7 4e-12
ath:AT3G55930 RNA splicing factor-related 54.3 2e-07
sce:YDR473C PRP3, RNA3; Prp3p; K12843 U4/U6 small nuclear ribo... 47.8 2e-05
ath:AT3G56790 RNA splicing factor-related 35.8 0.062
ath:AT4G34420 hypothetical protein 31.6 1.1
xla:398925 stk4, MGC68762, krs2, mst1, ysk3; serine/threonine ... 31.2 1.4
ath:AT2G01130 ATP binding / ATP-dependent helicase/ RNA bindin... 29.6 4.1
> tgo:TGME49_019790 U4/U6 small nuclear ribonucleoprotein, putative
; K12843 U4/U6 small nuclear ribonucleoprotein PRP3
Length=694
Score = 120 bits (302), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 81/126 (64%), Positives = 95/126 (75%), Gaps = 4/126 (3%)
Query 39 ARIWLSDCPAVEPWDLPLLKQTKTKAYEI--DETKIDALIEHPPPIATV--AKGEAIVNM 94
A+IW + P+VEPWD +LK ++ EI E +D L+EHP P+ V A I+NM
Sbjct 386 AKIWAAQPPSVEPWDACILKVDASRPDEILPQEDALDNLVEHPVPVRPVLDASTNVIINM 445
Query 95 YLTPAERKKLRRRKRQEREREKPDKIRMGLLPPPPPKCKLSNLMRVLGDVAVADPSKTEK 154
YLT ERKKLRRRKRQE+EREK DKIRMGL+PPPPPK KL+NLMRVLGD AVADPSK EK
Sbjct 446 YLTKQERKKLRRRKRQEKEREKQDKIRMGLMPPPPPKVKLTNLMRVLGDQAVADPSKVEK 505
Query 155 KVREQM 160
+VREQM
Sbjct 506 EVREQM 511
> dre:403072 prpf3, HPRP3, HPRP3P; PRP3 pre-mRNA processing factor
3 homolog (yeast); K12843 U4/U6 small nuclear ribonucleoprotein
PRP3
Length=700
Score = 100 bits (250), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 66/167 (39%), Positives = 94/167 (56%), Gaps = 16/167 (9%)
Query 4 QLQQQQQQAAEAAAGTDTKTEVSPQEKLETAAPDLARIWLSDCPAVEPWDLPLL-----K 58
QL++ Q + A+AA KT + KL AP + + P +E WD +L
Sbjct 366 QLEKLQMEIAQAA----KKTGIQASTKLALIAPK-KELGEGEIPNIEWWDSYILPFHINL 420
Query 59 QTKTKAYEIDETKIDALIEHP----PPIATVAKGEAIVNMYLTPAERKKLRRRKRQERER 114
T E++ + L+EHP PP+ T + +YLT E+KKLRR+ R+E ++
Sbjct 421 SAHTNLDEVEMHGVTNLVEHPTQMAPPVDT--DKPVTLGVYLTKKEQKKLRRQTRREGQK 478
Query 115 EKPDKIRMGLLPPPPPKCKLSNLMRVLGDVAVADPSKTEKKVREQMA 161
E +K+R+GL+PPP PK ++SNLMRVLG AV DP+K E VR QMA
Sbjct 479 EVQEKVRLGLMPPPEPKVRISNLMRVLGTEAVQDPTKVEAHVRAQMA 525
> xla:443589 prpf3, MGC130724; PRP3 pre-mRNA processing factor
3 homolog; K12843 U4/U6 small nuclear ribonucleoprotein PRP3
Length=679
Score = 97.1 bits (240), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 64/169 (37%), Positives = 94/169 (55%), Gaps = 16/169 (9%)
Query 2 QQQLQQQQQQAAEAAAGTDTKTEVSPQEKLETAAPDLARIWLSDCPAVEPWDLPLLKQTK 61
+ QL++ Q + A+AA KT + +L AP + + P +E WD ++
Sbjct 342 KSQLEKLQSEIAQAAK----KTGIQTSTRLAMIAPK-KELREGEIPEIEWWDSFIIPNGT 396
Query 62 TKAYEI-----DETKIDALIEHP----PPIATVAKGEAIVNMYLTPAERKKLRRRKRQER 112
E+ D I L+EHP PP+ A + +YLT E+KKLRR+ R+E
Sbjct 397 ELTEEVLLKREDFHGITNLVEHPAQLSPPVDKDAP--VTLGIYLTKKEQKKLRRQTRREG 454
Query 113 EREKPDKIRMGLLPPPPPKCKLSNLMRVLGDVAVADPSKTEKKVREQMA 161
++E +K+R+GL+PPP PK ++SNLMRVLG A+ DP+K E VR QMA
Sbjct 455 QKELQEKVRLGLMPPPEPKVRISNLMRVLGTEAIQDPTKVEAHVRAQMA 503
> hsa:9129 PRPF3, HPRP3, HPRP3P, PRP3, Prp3p, RP18; PRP3 pre-mRNA
processing factor 3 homolog (S. cerevisiae); K12843 U4/U6
small nuclear ribonucleoprotein PRP3
Length=683
Score = 95.1 bits (235), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 64/166 (38%), Positives = 92/166 (55%), Gaps = 15/166 (9%)
Query 4 QLQQQQQQAAEAAAGTDTKTEVSPQEKLETAAPDLARIWLSDCPAVEPWDLPLLKQ---- 59
QL++ Q + ++AA KT + +L AP + D P +E WD ++
Sbjct 347 QLEKLQAEISQAA----RKTGIHTSTRLALIAPK-KELKEGDIPEIEWWDSYIIPNGFDL 401
Query 60 TKTKAYEIDETKIDALIEHP----PPIATVAKGEAIVNMYLTPAERKKLRRRKRQERERE 115
T+ D I L+EHP PP+ + +YLT E+KKLRR+ R+E ++E
Sbjct 402 TEENPKREDYFGITNLVEHPAQLNPPVDN--DTPVTLGVYLTKKEQKKLRRQTRREAQKE 459
Query 116 KPDKIRMGLLPPPPPKCKLSNLMRVLGDVAVADPSKTEKKVREQMA 161
+K+R+GL+PPP PK ++SNLMRVLG AV DP+K E VR QMA
Sbjct 460 LQEKVRLGLMPPPEPKVRISNLMRVLGTEAVQDPTKVEAHVRAQMA 505
> mmu:70767 Prpf3, 3632413F13Rik; PRP3 pre-mRNA processing factor
3 homolog (yeast); K12843 U4/U6 small nuclear ribonucleoprotein
PRP3
Length=683
Score = 94.0 bits (232), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 64/166 (38%), Positives = 91/166 (54%), Gaps = 15/166 (9%)
Query 4 QLQQQQQQAAEAAAGTDTKTEVSPQEKLETAAPDLARIWLSDCPAVEPWDLPLLKQ---- 59
QL++ Q + ++AA KT + +L AP + D P +E WD ++
Sbjct 347 QLEKLQAEISQAA----RKTGIHTSTRLALIAPK-KELKEGDIPEIEWWDSYIIPNGFDL 401
Query 60 TKTKAYEIDETKIDALIEHP----PPIATVAKGEAIVNMYLTPAERKKLRRRKRQERERE 115
T+ D I L+EHP PP+ + +YLT E+KKLRR+ R+E ++E
Sbjct 402 TEENPKREDYFGITNLVEHPAQLNPPVDN--DTPVTLGVYLTKKEQKKLRRQTRREAQKE 459
Query 116 KPDKIRMGLLPPPPPKCKLSNLMRVLGDVAVADPSKTEKKVREQMA 161
+K+R+GL PPP PK ++SNLMRVLG AV DP+K E VR QMA
Sbjct 460 LQEKVRLGLTPPPEPKVRISNLMRVLGTEAVQDPTKVEAHVRAQMA 505
> cel:M03C11.7 hypothetical protein; K12843 U4/U6 small nuclear
ribonucleoprotein PRP3
Length=621
Score = 89.7 bits (221), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 59/166 (35%), Positives = 89/166 (53%), Gaps = 12/166 (7%)
Query 4 QLQQQQQQAAEAAAGTDTKTEVSPQEKLETAAPDLARIWLSDCPAVEPWDLPLLKQTKT- 62
+L++ Q + + AA T + V KL P + P +E WD+ +L +
Sbjct 280 KLERLQNEVSSAAQSTGISSAV----KLAMVTPTGTAKMENGVPDIEWWDMLVLDKVNYD 335
Query 63 -----KAYEIDETKIDALIEHPPPIA--TVAKGEAIVNMYLTPAERKKLRRRKRQERERE 115
E + L+EHP + T + + +YLT E+KK+RR+ R+E +E
Sbjct 336 EIPAENDMERYSQTVSELVEHPISMRAPTEPLTQQYLKVYLTTKEKKKIRRQNRKEVLKE 395
Query 116 KPDKIRMGLLPPPPPKCKLSNLMRVLGDVAVADPSKTEKKVREQMA 161
K +KIR+GL P PK K+SNLMRVLG+ A+ DP+K E +VR+QMA
Sbjct 396 KTEKIRLGLEKAPEPKVKISNLMRVLGNEAIQDPTKMEAQVRKQMA 441
> tpv:TP04_0348 hypothetical protein; K12843 U4/U6 small nuclear
ribonucleoprotein PRP3
Length=604
Score = 85.5 bits (210), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 54/140 (38%), Positives = 75/140 (53%), Gaps = 21/140 (15%)
Query 42 WLSDCPAVEPWDLPL-----------LKQTKTKAY---------EIDETKIDALIEHPPP 81
W D P VE WD + L+ K Y +I+E + + IEHP
Sbjct 295 WDDDEPFVEWWDRGVIILKSGEDPFKLQNLKKHQYITNVNLNELKINEDRFNNYIEHPVK 354
Query 82 I-ATVAKGEAIVNMYLTPAERKKLRRRKRQEREREKPDKIRMGLLPPPPPKCKLSNLMRV 140
+ KG IV LT ER+K R +R++R++E DKI +G +PPPPPK ++SNL+ V
Sbjct 355 LDGKRKKGLNIVPTQLTKKERRKARTLRRKQRQKEIRDKIAIGSIPPPPPKLRMSNLVNV 414
Query 141 LGDVAVADPSKTEKKVREQM 160
L + ADPSK EK V++QM
Sbjct 415 LKNQTAADPSKVEKMVKKQM 434
> cpv:cgd5_1740 conserved protein, possible U4/U6 associated RNA
splicing factor ; K12843 U4/U6 small nuclear ribonucleoprotein
PRP3
Length=452
Score = 84.0 bits (206), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 47/120 (39%), Positives = 72/120 (60%), Gaps = 7/120 (5%)
Query 47 PAVEPWDLPLLKQTKTKA-----YEIDETKIDALIEHPPPIATVAKGEAIVN--MYLTPA 99
P +E WD+P +K + + +EI +KI LIEHP P+ + V+ ++LT
Sbjct 175 PELEHWDMPFVKPKENVSEDEFPFEILVSKISNLIEHPVPLEPYFDPDENVSSVIFLTKK 234
Query 100 ERKKLRRRKRQEREREKPDKIRMGLLPPPPPKCKLSNLMRVLGDVAVADPSKTEKKVREQ 159
E+ +LRR RQE+E E+ D+IRMG+ P P KLSNL +L + VA+PS E ++++Q
Sbjct 235 EKARLRRMNRQEKEMERQDRIRMGIESPLRPILKLSNLHNILNEKVVAEPSNIELEIKKQ 294
> pfa:MAL13P1.45 U4/U6 small nuclear ribonucleoprotein, putative;
K12843 U4/U6 small nuclear ribonucleoprotein PRP3
Length=728
Score = 76.6 bits (187), Expect = 3e-14, Method: Composition-based stats.
Identities = 60/101 (59%), Positives = 76/101 (75%), Gaps = 3/101 (2%)
Query 63 KAYEIDETKIDALIEHPPPI---ATVAKGEAIVNMYLTPAERKKLRRRKRQEREREKPDK 119
+ Y I+ I + IEHP P+ + +++M+LTP ERKKLR+RKRQE+EREK DK
Sbjct 414 RLYSINLNCIKSYIEHPVPLNEDENYDDDKYLIHMFLTPQERKKLRKRKRQEKEREKQDK 473
Query 120 IRMGLLPPPPPKCKLSNLMRVLGDVAVADPSKTEKKVREQM 160
IR+GL+PPPPPK KLSNLMRV+GD A+A PSKTE VR+QM
Sbjct 474 IRIGLMPPPPPKMKLSNLMRVMGDTALAQPSKTEYAVRKQM 514
> bbo:BBOV_II002150 18.m06172; hypothetical protein; K12843 U4/U6
small nuclear ribonucleoprotein PRP3
Length=539
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 62/139 (44%), Positives = 81/139 (58%), Gaps = 20/139 (14%)
Query 42 WLSDCPAVEPWDLPLL---------KQTKTKAYE----------IDETKIDALIEHPPPI 82
W D P +E WD ++ K K ++ I+E K + IEHP I
Sbjct 226 WDDDEPWMEWWDRGVIILKTGEDPFKSGNVKKHQTLDIDPDLCMINEKKFNNYIEHPCKI 285
Query 83 -ATVAKGEAIVNMYLTPAERKKLRRRKRQEREREKPDKIRMGLLPPPPPKCKLSNLMRVL 141
KG+ ++ LT ERKKLRRRKR+E+ + DKIR+GL+PPP PK KLSNL+RVL
Sbjct 286 DGKRKKGKDTPSIMLTKEERKKLRRRKREEKLKAIQDKIRIGLIPPPAPKLKLSNLLRVL 345
Query 142 GDVAVADPSKTEKKVREQM 160
D +VADPS EK+VR+QM
Sbjct 346 KDQSVADPSLVEKQVRDQM 364
> ath:AT1G28060 small nuclear ribonucleoprotein family protein
/ snRNP family protein; K12843 U4/U6 small nuclear ribonucleoprotein
PRP3
Length=786
Score = 69.7 bits (169), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 49/124 (39%), Positives = 65/124 (52%), Gaps = 14/124 (11%)
Query 46 CPAVEPWDLPLLKQTKTKAYEIDET----------KIDALIEHP--PPIATVAKGEAIVN 93
P VE WD +L T + EI + K+ IEHP A
Sbjct 490 IPDVEWWDANVL--TNGEYGEITDGTITESHLKIEKLTHYIEHPRPIEPPAEAAPPPPQP 547
Query 94 MYLTPAERKKLRRRKRQEREREKPDKIRMGLLPPPPPKCKLSNLMRVLGDVAVADPSKTE 153
+ LT E+KKLR ++R +E+EK + IR GLL PP K K+SNLM+VLG A DP+K E
Sbjct 548 LKLTKKEQKKLRTQRRLAKEKEKQEMIRQGLLEPPKAKVKMSNLMKVLGSEATQDPTKLE 607
Query 154 KKVR 157
K++R
Sbjct 608 KEIR 611
> ath:AT3G55930 RNA splicing factor-related
Length=437
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 35/92 (38%), Positives = 51/92 (55%), Gaps = 2/92 (2%)
Query 72 IDALIEHPPPIATVAKGEAIVN--MYLTPAERKKLRRRKRQEREREKPDKIRMGLLPPPP 129
+ IEHP PI A+ + + +T ERKKLR +R +E EK + I G + P
Sbjct 181 FNCHIEHPFPIEPPAEAASPPPQPLKMTKEERKKLRTLRRVAKEMEKREMISQGRVEPQK 240
Query 130 PKCKLSNLMRVLGDVAVADPSKTEKKVREQMA 161
K K+SNLM+V A +P+K EK++R + A
Sbjct 241 SKVKMSNLMKVRASEATQNPTKLEKEIRTEAA 272
> sce:YDR473C PRP3, RNA3; Prp3p; K12843 U4/U6 small nuclear ribonucleoprotein
PRP3
Length=469
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 35/87 (40%), Positives = 52/87 (59%), Gaps = 2/87 (2%)
Query 75 LIEHPPPIATVAKGEAIVNMYLTPAERKKLRRRKRQEREREKPDKIRMGLLPPPPPKCKL 134
+ HP P + + + + YLT ERK+LRR +R+ + KI++GLLP P PK KL
Sbjct 216 YVAHPLP-EKINEAKVSIKAYLTQHERKRLRRNRRKMAREAREIKIKLGLLPKPEPKVKL 274
Query 135 SNLMRVL-GDVAVADPSKTEKKVREQM 160
SN+M V D + DP+ EK V++Q+
Sbjct 275 SNMMSVFENDQNITDPTAWEKVVKDQV 301
> ath:AT3G56790 RNA splicing factor-related
Length=206
Score = 35.8 bits (81), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 108 KRQEREREKPDKIRMGLLPPPPPKCKLSNLMRVLG 142
+R +E+EK IR L P K KLSNLM+VL
Sbjct 162 RRVAKEKEKQKMIRQALFEPQKSKVKLSNLMKVLA 196
> ath:AT4G34420 hypothetical protein
Length=491
Score = 31.6 bits (70), Expect = 1.1, Method: Composition-based stats.
Identities = 11/23 (47%), Positives = 14/23 (60%), Gaps = 0/23 (0%)
Query 47 PAVEPWDLPLLKQTKTKAYEIDE 69
P +EPWD+P + K K Y DE
Sbjct 218 PNIEPWDIPFVHYPKPKTYNRDE 240
> xla:398925 stk4, MGC68762, krs2, mst1, ysk3; serine/threonine
kinase 4 (EC:2.7.11.1); K04411 serine/threonine kinase 4 [EC:2.7.11.6]
Length=485
Score = 31.2 bits (69), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 49/97 (50%), Gaps = 13/97 (13%)
Query 75 LIEHPPPIATVAKGEAIVNMYLTPAERKKLRRRKRQERERE-------KPDKIRMGLLPP 127
L++HP AKGE+I+ L A+ +KL+R + ++RE E D++ +G +
Sbjct 275 LLQHP--FIKAAKGESILRHLLNAAQDEKLKRTELKQREVEPEEKENVNEDEVDVGTMVQ 332
Query 128 PPPKCKLSNLMRVLGDVA-VADPSKTEK-KVREQMAA 162
K N M+ LG ++ AD + EK K+ QM
Sbjct 333 AGGKD--LNTMKELGMMSEGADGTMVEKDKLETQMGT 367
> ath:AT2G01130 ATP binding / ATP-dependent helicase/ RNA binding
/ double-stranded RNA binding / helicase/ nucleic acid binding
Length=1113
Score = 29.6 bits (65), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 14/51 (27%), Positives = 28/51 (54%), Gaps = 0/51 (0%)
Query 72 IDALIEHPPPIATVAKGEAIVNMYLTPAERKKLRRRKRQEREREKPDKIRM 122
+ A++ PI TVA G ++ + +LTP ++K L + + R+ D + +
Sbjct 719 LGAILGCLDPILTVAAGLSVRDPFLTPQDKKDLAEAAKSQFSRDHSDHLAL 769
Lambda K H
0.312 0.128 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3767900632
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40