bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4595_orf2
Length=75
Score E
Sequences producing significant alignments: (Bits) Value
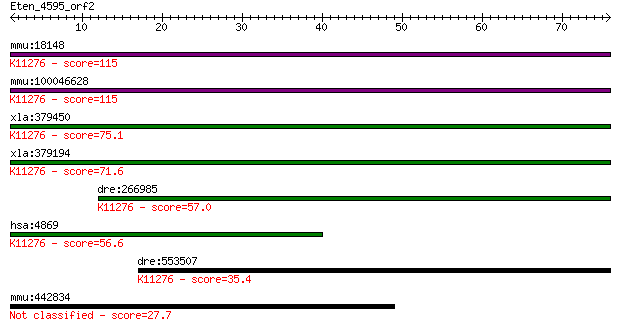
mmu:18148 Npm1, B23, MGC102162, MGC107291, NO38, Npm; nucleoph... 115 4e-26
mmu:100046628 nucleophosmin-like; K11276 nucleophosmin 1 115 4e-26
xla:379450 no38, MGC64305; nucleolar protein NO38; K11276 nucl... 75.1 5e-14
xla:379194 npm1, B23, MGC53901; nucleophosmin (nucleolar phosp... 71.6 6e-13
dre:266985 npm1, sb:cb487; nucleophosmin 1; K11276 nucleophosm... 57.0 2e-08
hsa:4869 NPM1, B23, MGC104254, NPM; nucleophosmin (nucleolar p... 56.6 2e-08
dre:553507 npm4; nucleophosmin/nucleoplasmin, 4; K11276 nucleo... 35.4 0.038
mmu:442834 D830031N03Rik, KIAA0754, mKIAA0754; RIKEN cDNA D830... 27.7 8.7
> mmu:18148 Npm1, B23, MGC102162, MGC107291, NO38, Npm; nucleophosmin
1; K11276 nucleophosmin 1
Length=292
Score = 115 bits (287), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 54/75 (72%), Positives = 59/75 (78%), Gaps = 0/75 (0%)
Query 1 PNSKGQNSFKNREKTPKPPKGPSFVEAFKAKMQPSLEKGGSLPKVEPKFFNFVKNFFRMP 60
P SKGQ SFK +EKTPK PKGPS VE KAKMQ S+EKGGSLPKVE KF N+VKN FRM
Sbjct 218 PRSKGQESFKKQEKTPKTPKGPSSVEDIKAKMQASIEKGGSLPKVEAKFINYVKNCFRMT 277
Query 61 AREAFQNFWQWRKFL 75
+EA Q+ WQWRK L
Sbjct 278 DQEAIQDLWQWRKSL 292
> mmu:100046628 nucleophosmin-like; K11276 nucleophosmin 1
Length=292
Score = 115 bits (287), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 54/75 (72%), Positives = 59/75 (78%), Gaps = 0/75 (0%)
Query 1 PNSKGQNSFKNREKTPKPPKGPSFVEAFKAKMQPSLEKGGSLPKVEPKFFNFVKNFFRMP 60
P SKGQ SFK +EKTPK PKGPS VE KAKMQ S+EKGGSLPKVE KF N+VKN FRM
Sbjct 218 PRSKGQESFKKQEKTPKTPKGPSSVEDIKAKMQASIEKGGSLPKVEAKFINYVKNCFRMT 277
Query 61 AREAFQNFWQWRKFL 75
+EA Q+ WQWRK L
Sbjct 278 DQEAIQDLWQWRKSL 292
> xla:379450 no38, MGC64305; nucleolar protein NO38; K11276 nucleophosmin
1
Length=299
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 38/75 (50%), Positives = 48/75 (64%), Gaps = 2/75 (2%)
Query 1 PNSKGQNSFKNREKTPKPPKGPSFVEAFKAKMQPSLEKGGSLPKVEPKFFNFVKNFFRMP 60
P KG+ K++ TPK PK P E KAKMQ LEKG LPKVE KF N+VKN FR
Sbjct 223 PEQKGKQDTKSQ--TPKTPKTPLSFEEMKAKMQTILEKGTVLPKVEVKFANYVKNCFRTD 280
Query 61 AREAFQNFWQWRKFL 75
+++ Q+ W+WR+ L
Sbjct 281 SQKVIQDLWKWRQSL 295
> xla:379194 npm1, B23, MGC53901; nucleophosmin (nucleolar phosphoprotein
B23, numatrin); K11276 nucleophosmin 1
Length=300
Score = 71.6 bits (174), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 37/75 (49%), Positives = 46/75 (61%), Gaps = 2/75 (2%)
Query 1 PNSKGQNSFKNREKTPKPPKGPSFVEAFKAKMQPSLEKGGSLPKVEPKFFNFVKNFFRMP 60
P KG+ K + TPK PK P E KAKMQ LEKG LPKVE KF N+VKN FR
Sbjct 224 PEQKGKQDTKPQ--TPKTPKTPLSSEEIKAKMQTYLEKGNVLPKVEVKFANYVKNCFRTE 281
Query 61 AREAFQNFWQWRKFL 75
++ ++ W+WR+ L
Sbjct 282 NQKVIEDLWKWRQSL 296
> dre:266985 npm1, sb:cb487; nucleophosmin 1; K11276 nucleophosmin
1
Length=282
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 40/64 (62%), Gaps = 0/64 (0%)
Query 12 REKTPKPPKGPSFVEAFKAKMQPSLEKGGSLPKVEPKFFNFVKNFFRMPAREAFQNFWQW 71
+E+TPK P+ P + K+KM S+ KG SLPKV+ KF N+V N F+ + + W+W
Sbjct 218 KEQTPKTPQTPRTLADIKSKMMESVAKGVSLPKVQLKFENYVNNCFKGTDPKVVEELWKW 277
Query 72 RKFL 75
R+ +
Sbjct 278 RQTV 281
> hsa:4869 NPM1, B23, MGC104254, NPM; nucleophosmin (nucleolar
phosphoprotein B23, numatrin); K11276 nucleophosmin 1
Length=259
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 28/39 (71%), Positives = 30/39 (76%), Gaps = 0/39 (0%)
Query 1 PNSKGQNSFKNREKTPKPPKGPSFVEAFKAKMQPSLEKG 39
P SKGQ SFK +EKTPK PKGPS VE KAKMQ S+EK
Sbjct 220 PRSKGQESFKKQEKTPKTPKGPSSVEDIKAKMQASIEKA 258
> dre:553507 npm4; nucleophosmin/nucleoplasmin, 4; K11276 nucleophosmin
1
Length=298
Score = 35.4 bits (80), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 33/59 (55%), Gaps = 1/59 (1%)
Query 17 KPPKGPSFVEAFKAKMQPSLEKGGSLPKVEPKFFNFVKNFFRMPAREAFQNFWQWRKFL 75
K P PS E K+K+ + ++G PK E KF NF ++ F++ ++ ++ W + + L
Sbjct 239 KTPSIPSLSE-VKSKLTSAAKEGKPFPKTEQKFENFARSSFKISDKQVIKDLWNFVQSL 296
> mmu:442834 D830031N03Rik, KIAA0754, mKIAA0754; RIKEN cDNA D830031N03
gene
Length=1115
Score = 27.7 bits (60), Expect = 8.7, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 25/49 (51%), Gaps = 1/49 (2%)
Query 1 PNSKGQNSFKNRE-KTPKPPKGPSFVEAFKAKMQPSLEKGGSLPKVEPK 48
PN++ +++ R K+ PP P F E +A + E G S P ++ K
Sbjct 120 PNAERKDNVNRRSWKSFMPPNFPEFAERMEASLSEVSEAGASNPSLQEK 168
Lambda K H
0.319 0.135 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2002740660
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40