bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
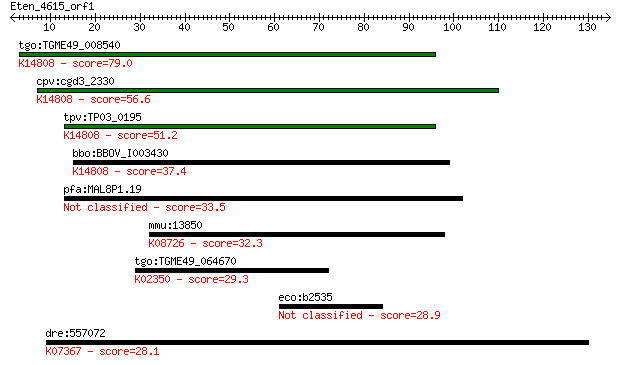
Query= Eten_4615_orf1
Length=134
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_008540 ATP-dependent RNA helicase, putative (EC:5.9... 79.0 4e-15
cpv:cgd3_2330 hypothetical protein ; K14808 ATP-dependent RNA ... 56.6 2e-08
tpv:TP03_0195 ATP-dependent RNA helicase; K14808 ATP-dependent... 51.2 1e-06
bbo:BBOV_I003430 19.m02227; DEAD/DEAH box helicase and helicas... 37.4 0.014
pfa:MAL8P1.19 RNA helicase, putative 33.5 0.20
mmu:13850 Ephx2, AW106936, Eph2, sEP; epoxide hydrolase 2, cyt... 32.3 0.42
tgo:TGME49_064670 DNA polymerase zeta catalytic subunit, putat... 29.3 3.2
eco:b2535 csiE, csi-16, ECK2532, JW5878; stationary phase indu... 28.9 4.9
dre:557072 FLJ00120 protein-like; K07367 caspase recruitment d... 28.1 6.8
> tgo:TGME49_008540 ATP-dependent RNA helicase, putative (EC:5.99.1.3);
K14808 ATP-dependent RNA helicase DDX54/DBP10 [EC:3.6.4.13]
Length=983
Score = 79.0 bits (193), Expect = 4e-15, Method: Composition-based stats.
Identities = 47/100 (47%), Positives = 57/100 (57%), Gaps = 7/100 (7%)
Query 3 EERKISCAFRYAQWCKRTKQRIQKVGEMENPNLNPPERSRSRASR-------TEEPPDTD 55
+E K S A RYAQWC+ TK+RIQ+VGE E+P L + + R + D D
Sbjct 852 QEEKTSSAHRYAQWCRETKRRIQRVGEEESPELAATPKDKRGKGRGADKHFVSSSDSDGD 911
Query 56 ESPSNKIRFLPLHSKHLGIKEALQKGAPLTHKQTRIAKKL 95
+ RFL KH GI+EALQKG LTHKQ RI KKL
Sbjct 912 GPGPSAGRFLAFTDKHKGIQEALQKGVKLTHKQKRIVKKL 951
> cpv:cgd3_2330 hypothetical protein ; K14808 ATP-dependent RNA
helicase DDX54/DBP10 [EC:3.6.4.13]
Length=862
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 38/104 (36%), Positives = 55/104 (52%), Gaps = 6/104 (5%)
Query 7 ISCAFRYAQWCKRTKQRIQKVGEMENPNLNPPERSRSRASRTEEPPDTDESPSNKIRFLP 66
+ C +Y +W T +IQ VGE+E+ LN R+ + D + S +
Sbjct 699 LKCTGQYKKWKANTNLKIQNVGEIEDTELNKTIRNNGFKDKYN-TSDINSS----FDYSN 753
Query 67 LHSKHLGIKEALQKGAPLTHKQTRIAKKL-IQKEINLNSKMNLQ 109
+H KH GI EA+ KG LTHKQ RIA+++ I KE +N +LQ
Sbjct 754 VHDKHKGIIEAISKGIKLTHKQKRIARRIGIIKEKIVNKSKSLQ 797
> tpv:TP03_0195 ATP-dependent RNA helicase; K14808 ATP-dependent
RNA helicase DDX54/DBP10 [EC:3.6.4.13]
Length=839
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 49/85 (57%), Gaps = 10/85 (11%)
Query 13 YAQWCKRTKQRIQKVGEMENPNLN--PPERSRSRASRTEEPPDTDESPSNKIRFLPLHSK 70
+ +W K+T +RIQ VGE+ENP+++ +R+ ++ S + + PD DE + + K
Sbjct 707 FKKWIKKTSRRIQHVGEVENPSISGFGADRTPTKNSLSCD-PDLDELKT-------MFPK 758
Query 71 HLGIKEALQKGAPLTHKQTRIAKKL 95
H I +A + LTHKQ RI KL
Sbjct 759 HKDIIDAYESNEQLTHKQIRILNKL 783
> bbo:BBOV_I003430 19.m02227; DEAD/DEAH box helicase and helicase
conserved C-terminal domain containing protein; K14808 ATP-dependent
RNA helicase DDX54/DBP10 [EC:3.6.4.13]
Length=783
Score = 37.4 bits (85), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 26/84 (30%), Positives = 38/84 (45%), Gaps = 3/84 (3%)
Query 15 QWCKRTKQRIQKVGEMENPNLNPPERSRSRASRTEEPPDTDESPSNKIRFLPLHSKHLGI 74
+W K +++RIQK+GE E P ++ + D S + P KH
Sbjct 647 KWMKTSRKRIQKLGEQEEYQAAPKKKKQMDEDAYSSGDDHKASYDELMHMFP---KHAAT 703
Query 75 KEALQKGAPLTHKQTRIAKKLIQK 98
EA + PLT+KQ R K+L K
Sbjct 704 LEAAKLSLPLTNKQKRTVKRLTTK 727
> pfa:MAL8P1.19 RNA helicase, putative
Length=1289
Score = 33.5 bits (75), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 34/110 (30%), Positives = 47/110 (42%), Gaps = 23/110 (20%)
Query 13 YAQWCKRTKQRIQKVGEMENPNLNPPERSRSRASRTEEPPDTDESPSNK----------- 61
Y +W K TK+RI+ VGE+E+ N P+ ++ + +++ +
Sbjct 1114 YEKWVKSTKKRIKNVGELEDENDIIPKTKLNKNKNKNKKGKKNKNQNKYNNDISDNNDNN 1173
Query 62 ---------IRFLPLHSKHLGIKEALQKGAPLTHKQTRIAKKLIQ-KEIN 101
I L L H I EAL K LT KQ RI KK I K IN
Sbjct 1174 SENEEKQYNINMLKL--MHPEISEALSKNNKLTKKQQRIYKKYISGKYIN 1221
> mmu:13850 Ephx2, AW106936, Eph2, sEP; epoxide hydrolase 2, cytoplasmic
(EC:3.3.2.10); K08726 soluble epoxide hydrolase [EC:3.3.2.10]
Length=554
Score = 32.3 bits (72), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 30/68 (44%), Gaps = 2/68 (2%)
Query 32 NPNLNPPERSRSRASRTE--EPPDTDESPSNKIRFLPLHSKHLGIKEALQKGAPLTHKQT 89
N L PER R+ AS PPD D SP IR +P+ + L +E A L +
Sbjct 341 NMALFYPERVRAVASLNTPFMPPDPDVSPMKVIRSIPVFNYQLYFQEPGVAEAELEKNMS 400
Query 90 RIAKKLIQ 97
R K +
Sbjct 401 RTFKSFFR 408
> tgo:TGME49_064670 DNA polymerase zeta catalytic subunit, putative
(EC:2.7.7.7); K02350 DNA polymerase zeta subunit [EC:2.7.7.7]
Length=3952
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 12/43 (27%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 29 EMENPNLNPPERSRSRASRTEEPPDTDESPSNKIRFLPLHSKH 71
E N PP+R++ + T PP D + N + + + S+H
Sbjct 1539 ESRNRQEAPPQRAKEKEEETASPPTPDRTRENTEKHVGMRSRH 1581
> eco:b2535 csiE, csi-16, ECK2532, JW5878; stationary phase inducible
protein
Length=426
Score = 28.9 bits (63), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 11/23 (47%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 61 KIRFLPLHSKHLGIKEALQKGAP 83
++ LPL+ KH+ +K LQ GAP
Sbjct 361 ELTLLPLNIKHMSVKAFLQTGAP 383
> dre:557072 FLJ00120 protein-like; K07367 caspase recruitment
domain-containing protein 11
Length=1357
Score = 28.1 bits (61), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 33/134 (24%), Positives = 55/134 (41%), Gaps = 16/134 (11%)
Query 9 CAFRYAQWCKRTKQRIQKVGEMENP--NLNPPERSR-------SRASR----TEEPPDTD 55
C+ R ++ Q +Q V ++N +N ER R S SR T P DT
Sbjct 1092 CSPRLSRASVFISQILQFVSRVDNKYKRMNSSERVRIVNGGNPSSVSRPGFETLRPEDTS 1151
Query 56 ESPSNKIRFLPLHSKHLGIKEALQKGAPLTHKQTRIAKKLIQKEINLNSKMNLQKGKKNN 115
+ S+ + L L L Q+ P+ +AK ++QK +NL M K
Sbjct 1152 DPESDLNKSLNLIPYSLVTPHQCQRKRPILFTPNILAKTIVQKFLNLGGAMEFTSWK--- 1208
Query 116 PEVYGQQQQQQQQQ 129
P++ + + +Q+
Sbjct 1209 PDIVTKDEFLMKQK 1222
Lambda K H
0.310 0.126 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2231140792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40