bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4636_orf2
Length=220
Score E
Sequences producing significant alignments: (Bits) Value
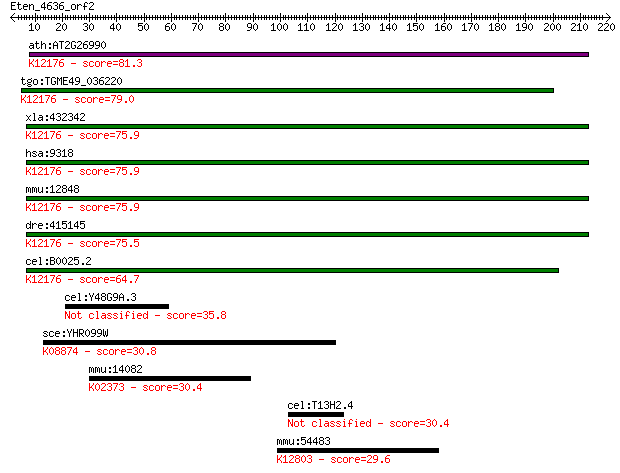
ath:AT2G26990 FUS12; FUS12 (FUSCA 12); K12176 COP9 signalosome... 81.3 3e-15
tgo:TGME49_036220 hypothetical protein ; K12176 COP9 signaloso... 79.0 1e-14
xla:432342 cops2, csn2; COP9 constitutive photomorphogenic hom... 75.9 9e-14
hsa:9318 COPS2, ALIEN, CSN2, SGN2, TRIP15; COP9 constitutive p... 75.9 1e-13
mmu:12848 Cops2, AI315723, C85265, Csn2, Sgn2, Trip15, alien-l... 75.9 1e-13
dre:415145 cops2, wu:fc05f05, wu:fc06g10, wu:fc60f04, wu:fl94f... 75.5 1e-13
cel:B0025.2 csn-2; COP-9 SigNalosome subunit family member (cs... 64.7 2e-10
cel:Y48G9A.3 hypothetical protein 35.8 0.12
sce:YHR099W TRA1; Subunit of SAGA and NuA4 histone acetyltrans... 30.8 3.8
mmu:14082 Fadd, Mort1/FADD; Fas (TNFRSF6)-associated via death... 30.4 4.5
cel:T13H2.4 pqn-65; Prion-like-(Q/N-rich)-domain-bearing prote... 30.4 5.0
mmu:54483 Mefv, FMF, MGC124344, MGC124345, TRIM20, pyrin; Medi... 29.6 8.0
> ath:AT2G26990 FUS12; FUS12 (FUSCA 12); K12176 COP9 signalosome
complex subunit 2
Length=439
Score = 81.3 bits (199), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 57/213 (26%), Positives = 97/213 (45%), Gaps = 32/213 (15%)
Query 8 QLLRLYMQLGEWTKAEDLLPGVHQA-----GRDPSLPPHQQLEAAAVEVLCCTYGRDWKL 62
+L ++ +GE+ + +L +H++ G D Q LE A+E+ T +D K
Sbjct 150 KLCNIWFDIGEYRRMTKILKELHKSCQKEDGTDDQKKGSQLLEVYAIEIQIYTETKDNKK 209
Query 63 LQQLLPHALRLSAASLDPRSAAAVRECGARLLIELHGRSDLQQRQQQQNDQQEPEEQHQP 122
L+QL AL + +A PR +RECG G+ + +RQ
Sbjct 210 LKQLYHKALAIKSAIPHPRIMGIIRECG--------GKMHMAERQ--------------- 246
Query 123 WTDIHAHLMAAFRHHQEIGDARAAAAALRRAVLSAPLAGSDVDPFSTREGKALQGDPDVQ 182
W + AF+++ E G+ R L+ VL+ L S+V+PF +E K + DP++
Sbjct 247 WEEAATDFFEAFKNYDEAGNQR-RIQCLKYLVLANMLMESEVNPFDGQEAKPYKNDPEIL 305
Query 183 SAKALRAAFEANDVAGVEKHLQVLESV---DPF 212
+ L AA++ N++ E+ L+ DPF
Sbjct 306 AMTNLIAAYQRNEIIEFERILKSNRRTIMDDPF 338
> tgo:TGME49_036220 hypothetical protein ; K12176 COP9 signalosome
complex subunit 2
Length=521
Score = 79.0 bits (193), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 70/239 (29%), Positives = 99/239 (41%), Gaps = 68/239 (28%)
Query 5 VCSQLLRLYMQLGEWTKAEDLLPGVHQAGR------------------------------ 34
CS+++RLY+ GE+ KA LL V + R
Sbjct 207 TCSRMIRLYIHQGEFPKATTLLADVRREARLTPLDISDYQLLSSSRSSLFPTPPSSSSSS 266
Query 35 ------------DPSLPPHQQLEAAAVEVLCCTYGRDWKLLQQLLPHALRLSAASL-DPR 81
+P Q LE A+E + C R++ L++L A + A + DP
Sbjct 267 SSSAFASAGSGESEEMPSGQILEFYALESVVCMRQRNFPRLRRLAAEAEKFLLAGIADPT 326
Query 82 SAAAVRECGARL-LIELHGRSDLQQRQQQQNDQQEPEEQHQPWTDIHAHLMAAFRHHQEI 140
A VRE ++ ++E + W AFRH QE+
Sbjct 327 HVATVREITGKIHMVE------------------------KRWKRALVDFSEAFRHFQEV 362
Query 141 GDARAAAAALRRAVLSAPLAGSDVDPFSTREGKALQGDPDVQSAKALRAAFEANDVAGV 199
G + A LR VL++ L+ SD+ PF TRE KALQGDP V + ALR A+EA+DV V
Sbjct 363 GRSTKAKQMLRLLVLASLLSLSDISPFDTREAKALQGDPSVAAMHALRRAYEADDVGRV 421
> xla:432342 cops2, csn2; COP9 constitutive photomorphogenic homolog
subunit 2; K12176 COP9 signalosome complex subunit 2
Length=443
Score = 75.9 bits (185), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 58/214 (27%), Positives = 98/214 (45%), Gaps = 32/214 (14%)
Query 7 SQLLRLYMQLGEWTKAEDLLPGVHQA-----GRDPSLPPHQQLEAAAVEVLCCTYGRDWK 61
++L +LY++ E+ K + +L +HQ+ G D Q LE A+E+ T ++ K
Sbjct 153 TKLGKLYLEREEYGKLQKILRQLHQSCQTDDGEDDLKKGTQLLEIYALEIQMYTAQKNNK 212
Query 62 LLQQLLPHALRLSAASLDPRSAAAVRECGARLLIELHGRSDLQQRQQQQNDQQEPEEQHQ 121
L+ L +L + +A P +RECG ++ H R E E
Sbjct 213 KLKALYEQSLHIKSAIPHPLIMGVIRECGGKM----HLR--------------EGE---- 250
Query 122 PWTDIHAHLMAAFRHHQEIGDARAAAAALRRAVLSAPLAGSDVDPFSTREGKALQGDPDV 181
+ H AF+++ E G R L+ VL+ L S ++PF ++E K + DP++
Sbjct 251 -FEKAHTDFFEAFKNYDESGSPRRTTC-LKYLVLANMLMKSGINPFDSQEAKPYKNDPEI 308
Query 182 QSAKALRAAFEANDVAGVEKHLQVLESV---DPF 212
+ L +A++ ND+ EK L+ S DPF
Sbjct 309 LAMTNLVSAYQNNDITEFEKILKTNHSNIMDDPF 342
> hsa:9318 COPS2, ALIEN, CSN2, SGN2, TRIP15; COP9 constitutive
photomorphogenic homolog subunit 2 (Arabidopsis); K12176 COP9
signalosome complex subunit 2
Length=450
Score = 75.9 bits (185), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 58/214 (27%), Positives = 98/214 (45%), Gaps = 32/214 (14%)
Query 7 SQLLRLYMQLGEWTKAEDLLPGVHQA-----GRDPSLPPHQQLEAAAVEVLCCTYGRDWK 61
++L +LY++ E+ K + +L +HQ+ G D Q LE A+E+ T ++ K
Sbjct 160 TKLGKLYLEREEYGKLQKILRQLHQSCQTDDGEDDLKKGTQLLEIYALEIQMYTAQKNNK 219
Query 62 LLQQLLPHALRLSAASLDPRSAAAVRECGARLLIELHGRSDLQQRQQQQNDQQEPEEQHQ 121
L+ L +L + +A P +RECG ++ H R E E
Sbjct 220 KLKALYEQSLHIKSAIPHPLIMGVIRECGGKM----HLR--------------EGE---- 257
Query 122 PWTDIHAHLMAAFRHHQEIGDARAAAAALRRAVLSAPLAGSDVDPFSTREGKALQGDPDV 181
+ H AF+++ E G R L+ VL+ L S ++PF ++E K + DP++
Sbjct 258 -FEKAHTDFFEAFKNYDESGSPRRTTC-LKYLVLANMLMKSGINPFDSQEAKPYKNDPEI 315
Query 182 QSAKALRAAFEANDVAGVEKHLQVLESV---DPF 212
+ L +A++ ND+ EK L+ S DPF
Sbjct 316 LAMTNLVSAYQNNDITEFEKILKTNHSNIMDDPF 349
> mmu:12848 Cops2, AI315723, C85265, Csn2, Sgn2, Trip15, alien-like;
COP9 (constitutive photomorphogenic) homolog, subunit
2 (Arabidopsis thaliana); K12176 COP9 signalosome complex subunit
2
Length=443
Score = 75.9 bits (185), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 58/214 (27%), Positives = 98/214 (45%), Gaps = 32/214 (14%)
Query 7 SQLLRLYMQLGEWTKAEDLLPGVHQA-----GRDPSLPPHQQLEAAAVEVLCCTYGRDWK 61
++L +LY++ E+ K + +L +HQ+ G D Q LE A+E+ T ++ K
Sbjct 153 TKLGKLYLEREEYGKLQKILRQLHQSCQTDDGEDDLKKGTQLLEIYALEIQMYTAQKNNK 212
Query 62 LLQQLLPHALRLSAASLDPRSAAAVRECGARLLIELHGRSDLQQRQQQQNDQQEPEEQHQ 121
L+ L +L + +A P +RECG ++ H R E E
Sbjct 213 KLKALYEQSLHIKSAIPHPLIMGVIRECGGKM----HLR--------------EGE---- 250
Query 122 PWTDIHAHLMAAFRHHQEIGDARAAAAALRRAVLSAPLAGSDVDPFSTREGKALQGDPDV 181
+ H AF+++ E G R L+ VL+ L S ++PF ++E K + DP++
Sbjct 251 -FEKAHTDFFEAFKNYDESGSPRRTTC-LKYLVLANMLMKSGINPFDSQEAKPYKNDPEI 308
Query 182 QSAKALRAAFEANDVAGVEKHLQVLESV---DPF 212
+ L +A++ ND+ EK L+ S DPF
Sbjct 309 LAMTNLVSAYQNNDITEFEKILKTNHSNIMDDPF 342
> dre:415145 cops2, wu:fc05f05, wu:fc06g10, wu:fc60f04, wu:fl94f12,
zgc:86624; COP9 constitutive photomorphogenic homolog
subunit 2 (Arabidopsis); K12176 COP9 signalosome complex subunit
2
Length=443
Score = 75.5 bits (184), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 58/214 (27%), Positives = 98/214 (45%), Gaps = 32/214 (14%)
Query 7 SQLLRLYMQLGEWTKAEDLLPGVHQA-----GRDPSLPPHQQLEAAAVEVLCCTYGRDWK 61
++L +LY++ E+ K + +L +HQ+ G D Q LE A+E+ T ++ K
Sbjct 153 TKLGKLYLEREEFGKLQKILRQLHQSCQTDDGEDDLKKGTQLLEIYALEIQMYTAQKNNK 212
Query 62 LLQQLLPHALRLSAASLDPRSAAAVRECGARLLIELHGRSDLQQRQQQQNDQQEPEEQHQ 121
L+ L +L + +A P +RECG ++ H R E E
Sbjct 213 KLKALYEQSLHIKSAIPHPLIMGVIRECGGKM----HLR--------------EGE---- 250
Query 122 PWTDIHAHLMAAFRHHQEIGDARAAAAALRRAVLSAPLAGSDVDPFSTREGKALQGDPDV 181
+ H AF+++ E G R L+ VL+ L S ++PF ++E K + DP++
Sbjct 251 -FEKAHTDFFEAFKNYDESGSPRRTTC-LKYLVLANMLMKSGINPFDSQEAKPYKNDPEI 308
Query 182 QSAKALRAAFEANDVAGVEKHLQVLESV---DPF 212
+ L +A++ ND+ EK L+ S DPF
Sbjct 309 LAMTNLVSAYQNNDITEFEKILKTNHSNIMDDPF 342
> cel:B0025.2 csn-2; COP-9 SigNalosome subunit family member (csn-2);
K12176 COP9 signalosome complex subunit 2
Length=495
Score = 64.7 bits (156), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 48/203 (23%), Positives = 91/203 (44%), Gaps = 32/203 (15%)
Query 7 SQLLRLYMQLGEWTKAEDLLPGV-----HQAGRDPSLPPHQQLEAAAVEVLCCTYGRDWK 61
++L +L+ L E+TK E ++ + ++ G + Q LE A+E+ T ++ K
Sbjct 150 TKLGKLFFDLHEFTKLEKIVKQLKVSCKNEQGEEDQRKGTQLLEIYALEIQMYTEQKNNK 209
Query 62 LLQ---QLLPHALRLSAASLDPRSAAAVRECGARLLIELHGRSDLQQRQQQQNDQQEPEE 118
L+ +L A+ +A P +RECG ++ + GR
Sbjct 210 ALKWVYELATQAIHTKSAIPHPLILGTIRECGGKMHLR-DGR------------------ 250
Query 119 QHQPWTDIHAHLMAAFRHHQEIGDARAAAAALRRAVLSAPLAGSDVDPFSTREGKALQGD 178
+ D H AF+++ E G R L+ VL+ L SD++PF ++E K + +
Sbjct 251 ----FLDAHTDFFEAFKNYDESGSPRRTTC-LKYLVLANMLIKSDINPFDSQEAKPFKNE 305
Query 179 PDVQSAKALRAAFEANDVAGVEK 201
P++ + + A++ ND+ E+
Sbjct 306 PEIVAMTQMVQAYQDNDIQAFEQ 328
> cel:Y48G9A.3 hypothetical protein
Length=2680
Score = 35.8 bits (81), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 21 KAEDLLPGVHQAGRDPSLPPHQQLEAAAVEVLCCTYGR 58
K DLL +H+ D P H++ A+E+LCCT G+
Sbjct 1429 KDTDLLGSIHKNMEDKKSPKHREGGLLALEILCCTIGK 1466
> sce:YHR099W TRA1; Subunit of SAGA and NuA4 histone acetyltransferase
complexes; interacts with acidic activators (e.g.,
Gal4p) which leads to transcription activation; similar to human
TRRAP, which is a cofactor for C-Myc mediated oncogenic
transformation; K08874 transformation/transcription domain-associated
protein
Length=3744
Score = 30.8 bits (68), Expect = 3.8, Method: Composition-based stats.
Identities = 29/110 (26%), Positives = 41/110 (37%), Gaps = 14/110 (12%)
Query 13 YMQLGEWTKAEDLLPGVHQAGRDPSLPPHQQLEAAAVE---VLCCTYGRDWKLLQQLLPH 69
Y Q+G W KA+ L R +L P+ Q E A E + C + W +L +L H
Sbjct 2694 YEQIGLWDKAQQLYEVAQVKARSGAL-PYSQSEYALWEDNWIQCAEKLQHWDVLTELAKH 2752
Query 70 ALRLSAASLDPRSAAAVRECGARLLIELHGRSDLQQRQQQQNDQQEPEEQ 119
+ ECG R+ R L+Q + D P Q
Sbjct 2753 E----------GFTDLLLECGWRVADWNSDRDALEQSVKSVMDVPTPRRQ 2792
> mmu:14082 Fadd, Mort1/FADD; Fas (TNFRSF6)-associated via death
domain; K02373 FAS (TNFRSF6)-associated via death domain
Length=205
Score = 30.4 bits (67), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 32/66 (48%), Gaps = 11/66 (16%)
Query 30 HQAGRDPSLPPHQQLEAAAVEVLCCTYGRDWKLLQQLLPHALRLSAASLD------PRSA 83
+AG + PP + A +++C GRDWK L L++S A +D PRS
Sbjct 82 FEAGTATAAPPGEADLQVAFDIVCDNVGRDWK----RLARELKVSEAKMDGIEEKYPRSL 137
Query 84 AA-VRE 88
+ VRE
Sbjct 138 SERVRE 143
> cel:T13H2.4 pqn-65; Prion-like-(Q/N-rich)-domain-bearing protein
family member (pqn-65)
Length=1702
Score = 30.4 bits (67), Expect = 5.0, Method: Composition-based stats.
Identities = 12/20 (60%), Positives = 14/20 (70%), Gaps = 0/20 (0%)
Query 103 LQQRQQQQNDQQEPEEQHQP 122
LQ + Q N QQ+PE QHQP
Sbjct 1160 LQDHEHQYNSQQQPEYQHQP 1179
> mmu:54483 Mefv, FMF, MGC124344, MGC124345, TRIM20, pyrin; Mediterranean
fever; K12803 pyrin
Length=808
Score = 29.6 bits (65), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 32/63 (50%), Gaps = 4/63 (6%)
Query 99 GRSDLQQRQQQQNDQQEPEE---QHQPWTDIHAHLMAAFRHHQEIGDARAAAAA-LRRAV 154
G+ ++ ++ DQ+ PE Q +PWT A L + Q GD + A+A LRR V
Sbjct 145 GKGPQKKSLTKRKDQRGPESLDSQTKPWTRSTAPLYRRTQGTQSPGDKESTASAQLRRNV 204
Query 155 LSA 157
SA
Sbjct 205 SSA 207
Lambda K H
0.319 0.132 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7202251840
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40