bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4641_orf2
Length=115
Score E
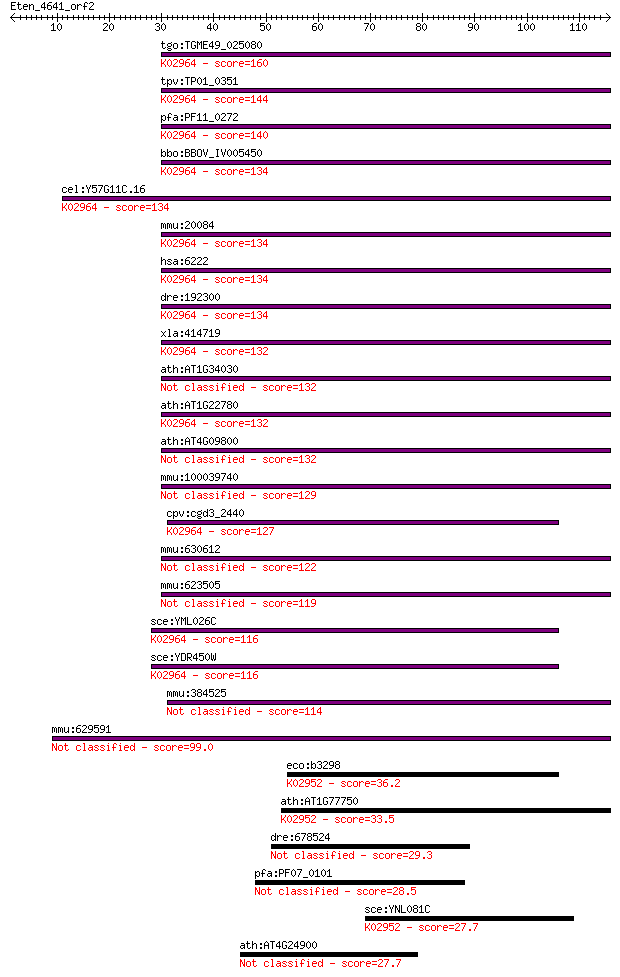
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_025080 40S ribosomal protein S18, putative ; K02964... 160 1e-39
tpv:TP01_0351 40S ribosomal protein S18; K02964 small subunit ... 144 7e-35
pfa:PF11_0272 40S ribosomal protein S18, putative; K02964 smal... 140 8e-34
bbo:BBOV_IV005450 23.m06078; 40S ribosomal protein S18; K02964... 134 5e-32
cel:Y57G11C.16 rps-18; Ribosomal Protein, Small subunit family... 134 8e-32
mmu:20084 Rps18, H-2Ke3, H2-Ke3, Ke-3; ribosomal protein S18; ... 134 8e-32
hsa:6222 RPS18, D6S218E, HKE3, KE-3, KE3, MGC117351, MGC126835... 134 8e-32
dre:192300 rps18, chunp6895, ke3; ribosomal protein S18; K0296... 134 8e-32
xla:414719 rps18, MGC82306; ribosomal protein S18; K02964 smal... 132 2e-31
ath:AT1G34030 40S ribosomal protein S18 (RPS18B) 132 3e-31
ath:AT1G22780 PFL; PFL (POINTED FIRST LEAVES); RNA binding / n... 132 3e-31
ath:AT4G09800 RPS18C; RPS18C (S18 RIBOSOMAL PROTEIN); RNA bind... 132 3e-31
mmu:100039740 Gm10260; ribosomal protein S18 pseudogene 129 2e-30
cpv:cgd3_2440 ribosomal protein S18A, rps18ap, HhH domain ; K0... 127 1e-29
mmu:630612 predicted gene, EG630612 122 2e-28
mmu:623505 Gm6436, EG623505; predicted gene 6436 119 3e-27
sce:YML026C RPS18B; Rps18bp; K02964 small subunit ribosomal pr... 116 1e-26
sce:YDR450W RPS18A; Rps18ap; K02964 small subunit ribosomal pr... 116 1e-26
mmu:384525 Gm5321, EG384525; predicted gene 5321 114 5e-26
mmu:629591 Gm6987, EG629591; predicted pseudogene 6987 99.0
eco:b3298 rpsM, ECK3285, JW3260; 30S ribosomal subunit protein... 36.2 0.027
ath:AT1G77750 30S ribosomal protein S13, chloroplast, putative... 33.5 0.15
dre:678524 MGC136872; zgc:136872 29.3 2.8
pfa:PF07_0101 conserved Plasmodium protein, unknown function 28.5 5.6
sce:YNL081C SWS2; Putative mitochondrial ribosomal protein of ... 27.7 8.3
ath:AT4G24900 hypothetical protein 27.7 8.5
> tgo:TGME49_025080 40S ribosomal protein S18, putative ; K02964
small subunit ribosomal protein S18e
Length=156
Score = 160 bits (404), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 72/86 (83%), Positives = 79/86 (91%), Gaps = 0/86 (0%)
Query 30 QIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRH 89
++VAI ++P QF IP+WFLNRQKDVKDGK +HV AN LD+VWRED ERLKKMRLHRGLRH
Sbjct 68 KVVAIASTPQQFKIPTWFLNRQKDVKDGKHKHVVANGLDTVWREDLERLKKMRLHRGLRH 127
Query 90 YWGLKVRGQHTKTTGRHGRTVGVAKK 115
YWGLKVRGQHTKTTGRHGRTVGVAKK
Sbjct 128 YWGLKVRGQHTKTTGRHGRTVGVAKK 153
> tpv:TP01_0351 40S ribosomal protein S18; K02964 small subunit
ribosomal protein S18e
Length=154
Score = 144 bits (363), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 65/86 (75%), Positives = 75/86 (87%), Gaps = 0/86 (0%)
Query 30 QIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRH 89
++VAI++SP +F +P WFLNRQKDVK+GK H ++N LDS RED ERLKKMRLHRGLRH
Sbjct 68 KLVAIISSPAEFKVPHWFLNRQKDVKEGKHMHNASNLLDSCLREDLERLKKMRLHRGLRH 127
Query 90 YWGLKVRGQHTKTTGRHGRTVGVAKK 115
YWGL+ RGQHTKTTGRHGRTVGVAKK
Sbjct 128 YWGLRTRGQHTKTTGRHGRTVGVAKK 153
> pfa:PF11_0272 40S ribosomal protein S18, putative; K02964 small
subunit ribosomal protein S18e
Length=156
Score = 140 bits (354), Expect = 8e-34, Method: Compositional matrix adjust.
Identities = 64/86 (74%), Positives = 75/86 (87%), Gaps = 0/86 (0%)
Query 30 QIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRH 89
IV I+++PTQF IP WFLNR+KD+K+GK+ HV AN LDS RED ER+KK+RLHRGLRH
Sbjct 68 NIVHIMSTPTQFKIPDWFLNRRKDLKEGKNIHVIANQLDSYLREDLERMKKIRLHRGLRH 127
Query 90 YWGLKVRGQHTKTTGRHGRTVGVAKK 115
+WGL+VRGQHTKTTGR GRTVGVAKK
Sbjct 128 HWGLRVRGQHTKTTGRRGRTVGVAKK 153
> bbo:BBOV_IV005450 23.m06078; 40S ribosomal protein S18; K02964
small subunit ribosomal protein S18e
Length=154
Score = 134 bits (338), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 58/86 (67%), Positives = 72/86 (83%), Gaps = 0/86 (0%)
Query 30 QIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRH 89
++V I++SP IP+WFLNRQKD K+GK+ H +AN LD+ R+D ER+KKMRLHRGLRH
Sbjct 68 KVVTIISSPADVMIPAWFLNRQKDYKEGKNLHNTANMLDTCLRDDLERMKKMRLHRGLRH 127
Query 90 YWGLKVRGQHTKTTGRHGRTVGVAKK 115
YWGL+ RGQHTK+TGRHG+TVGV KK
Sbjct 128 YWGLRTRGQHTKSTGRHGKTVGVVKK 153
> cel:Y57G11C.16 rps-18; Ribosomal Protein, Small subunit family
member (rps-18); K02964 small subunit ribosomal protein S18e
Length=154
Score = 134 bits (336), Expect = 8e-32, Method: Compositional matrix adjust.
Identities = 64/111 (57%), Positives = 80/111 (72%), Gaps = 6/111 (5%)
Query 11 AHCCCSPAA------AALLLLLLLLQIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSA 64
A CC A A L +IV I+ +P+Q+ IP+WFLNRQKD+KDGK+ + +
Sbjct 41 AFVCCRKADVDVNKRAGELTEEDFDKIVTIMQNPSQYKIPNWFLNRQKDIKDGKTGQLLS 100
Query 65 NNLDSVWREDFERLKKMRLHRGLRHYWGLKVRGQHTKTTGRHGRTVGVAKK 115
+D+ RED ER+KK+RLHRGLRHYWGL+VRGQHTKTTGR GRTVGV+KK
Sbjct 101 TAVDNKLREDLERMKKIRLHRGLRHYWGLRVRGQHTKTTGRKGRTVGVSKK 151
> mmu:20084 Rps18, H-2Ke3, H2-Ke3, Ke-3; ribosomal protein S18;
K02964 small subunit ribosomal protein S18e
Length=152
Score = 134 bits (336), Expect = 8e-32, Method: Compositional matrix adjust.
Identities = 60/86 (69%), Positives = 71/86 (82%), Gaps = 0/86 (0%)
Query 30 QIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRH 89
+++ I+ +P Q+ IP WFLNRQKDVKDGK V AN LD+ RED ERLKK+R HRGLRH
Sbjct 66 RVITIMQNPRQYKIPDWFLNRQKDVKDGKYSQVLANGLDNKLREDLERLKKIRAHRGLRH 125
Query 90 YWGLKVRGQHTKTTGRHGRTVGVAKK 115
+WGL+VRGQHTKTTGR GRTVGV+KK
Sbjct 126 FWGLRVRGQHTKTTGRRGRTVGVSKK 151
> hsa:6222 RPS18, D6S218E, HKE3, KE-3, KE3, MGC117351, MGC126835,
MGC126837; ribosomal protein S18; K02964 small subunit ribosomal
protein S18e
Length=152
Score = 134 bits (336), Expect = 8e-32, Method: Compositional matrix adjust.
Identities = 60/86 (69%), Positives = 71/86 (82%), Gaps = 0/86 (0%)
Query 30 QIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRH 89
+++ I+ +P Q+ IP WFLNRQKDVKDGK V AN LD+ RED ERLKK+R HRGLRH
Sbjct 66 RVITIMQNPRQYKIPDWFLNRQKDVKDGKYSQVLANGLDNKLREDLERLKKIRAHRGLRH 125
Query 90 YWGLKVRGQHTKTTGRHGRTVGVAKK 115
+WGL+VRGQHTKTTGR GRTVGV+KK
Sbjct 126 FWGLRVRGQHTKTTGRRGRTVGVSKK 151
> dre:192300 rps18, chunp6895, ke3; ribosomal protein S18; K02964
small subunit ribosomal protein S18e
Length=152
Score = 134 bits (336), Expect = 8e-32, Method: Compositional matrix adjust.
Identities = 60/86 (69%), Positives = 71/86 (82%), Gaps = 0/86 (0%)
Query 30 QIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRH 89
++V I+ +P Q+ IP WFLNRQKD+KDGK V AN LD+ RED ERLKK+R HRGLRH
Sbjct 66 RVVTIMQNPRQYKIPDWFLNRQKDIKDGKYSQVLANGLDNKLREDLERLKKIRAHRGLRH 125
Query 90 YWGLKVRGQHTKTTGRHGRTVGVAKK 115
+WGL+VRGQHTKTTGR GRTVGV+KK
Sbjct 126 FWGLRVRGQHTKTTGRRGRTVGVSKK 151
> xla:414719 rps18, MGC82306; ribosomal protein S18; K02964 small
subunit ribosomal protein S18e
Length=152
Score = 132 bits (333), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 59/86 (68%), Positives = 71/86 (82%), Gaps = 0/86 (0%)
Query 30 QIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRH 89
++V I+ +P Q+ IP WFLNRQKD+KDGK V AN LD+ RED ERLKK++ HRGLRH
Sbjct 66 RVVTIMQNPRQYKIPDWFLNRQKDIKDGKYSQVLANGLDNKLREDLERLKKIKAHRGLRH 125
Query 90 YWGLKVRGQHTKTTGRHGRTVGVAKK 115
+WGL+VRGQHTKTTGR GRTVGV+KK
Sbjct 126 FWGLRVRGQHTKTTGRRGRTVGVSKK 151
> ath:AT1G34030 40S ribosomal protein S18 (RPS18B)
Length=152
Score = 132 bits (331), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 60/86 (69%), Positives = 69/86 (80%), Gaps = 0/86 (0%)
Query 30 QIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRH 89
++ IVA+P QF IP WFLNRQKD KDGK V +N LD R+D ERLKK+R HRGLRH
Sbjct 66 NLMTIVANPRQFKIPDWFLNRQKDYKDGKYSQVVSNALDMKLRDDLERLKKIRNHRGLRH 125
Query 90 YWGLKVRGQHTKTTGRHGRTVGVAKK 115
YWGL+VRGQHTKTTGR G+TVGV+KK
Sbjct 126 YWGLRVRGQHTKTTGRRGKTVGVSKK 151
> ath:AT1G22780 PFL; PFL (POINTED FIRST LEAVES); RNA binding /
nucleic acid binding / structural constituent of ribosome;
K02964 small subunit ribosomal protein S18e
Length=152
Score = 132 bits (331), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 60/86 (69%), Positives = 69/86 (80%), Gaps = 0/86 (0%)
Query 30 QIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRH 89
++ IVA+P QF IP WFLNRQKD KDGK V +N LD R+D ERLKK+R HRGLRH
Sbjct 66 NLMTIVANPRQFKIPDWFLNRQKDYKDGKYSQVVSNALDMKLRDDLERLKKIRNHRGLRH 125
Query 90 YWGLKVRGQHTKTTGRHGRTVGVAKK 115
YWGL+VRGQHTKTTGR G+TVGV+KK
Sbjct 126 YWGLRVRGQHTKTTGRRGKTVGVSKK 151
> ath:AT4G09800 RPS18C; RPS18C (S18 RIBOSOMAL PROTEIN); RNA binding
/ nucleic acid binding / structural constituent of ribosome
Length=152
Score = 132 bits (331), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 60/86 (69%), Positives = 69/86 (80%), Gaps = 0/86 (0%)
Query 30 QIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRH 89
++ IVA+P QF IP WFLNRQKD KDGK V +N LD R+D ERLKK+R HRGLRH
Sbjct 66 NLMTIVANPRQFKIPDWFLNRQKDYKDGKYSQVVSNALDMKLRDDLERLKKIRNHRGLRH 125
Query 90 YWGLKVRGQHTKTTGRHGRTVGVAKK 115
YWGL+VRGQHTKTTGR G+TVGV+KK
Sbjct 126 YWGLRVRGQHTKTTGRRGKTVGVSKK 151
> mmu:100039740 Gm10260; ribosomal protein S18 pseudogene
Length=152
Score = 129 bits (325), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 58/86 (67%), Positives = 70/86 (81%), Gaps = 0/86 (0%)
Query 30 QIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRH 89
+++ I+ +P Q+ IP WFLNRQKDVKDGK V AN LD+ RED ERLKK+R HRGL H
Sbjct 66 RVITIMQNPRQYKIPDWFLNRQKDVKDGKYSQVLANGLDNKLREDLERLKKIRAHRGLHH 125
Query 90 YWGLKVRGQHTKTTGRHGRTVGVAKK 115
+WGL+VRGQHTKTTGR G+TVGV+KK
Sbjct 126 FWGLRVRGQHTKTTGRRGQTVGVSKK 151
> cpv:cgd3_2440 ribosomal protein S18A, rps18ap, HhH domain ;
K02964 small subunit ribosomal protein S18e
Length=153
Score = 127 bits (318), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 55/75 (73%), Positives = 68/75 (90%), Gaps = 0/75 (0%)
Query 31 IVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRHY 90
IVAIVA+P QF+IPS+ LNRQKDVKDGK +H+++NNL++ RED ERLKK+R HRGLRH+
Sbjct 68 IVAIVANPLQFNIPSYMLNRQKDVKDGKYKHITSNNLEASLREDLERLKKIRSHRGLRHH 127
Query 91 WGLKVRGQHTKTTGR 105
WG++VRGQHTKTTGR
Sbjct 128 WGVRVRGQHTKTTGR 142
> mmu:630612 predicted gene, EG630612
Length=166
Score = 122 bits (307), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 55/86 (63%), Positives = 69/86 (80%), Gaps = 0/86 (0%)
Query 30 QIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRH 89
+++ I+ +P Q++IP WFLNRQKDVKDGK + AN D+ ED ERLKK+R HR LRH
Sbjct 80 RVITIMQNPRQYNIPDWFLNRQKDVKDGKCSQILANGPDNKLPEDPERLKKIRDHRELRH 139
Query 90 YWGLKVRGQHTKTTGRHGRTVGVAKK 115
+WGL+VRGQHTKTTGR GRT+GV+KK
Sbjct 140 FWGLRVRGQHTKTTGRRGRTMGVSKK 165
> mmu:623505 Gm6436, EG623505; predicted gene 6436
Length=186
Score = 119 bits (297), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 54/86 (62%), Positives = 67/86 (77%), Gaps = 0/86 (0%)
Query 30 QIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRH 89
+++ I+ +P Q+SIP WFLNRQK VKDGK V A LD+ ED ERLKK+R HR L H
Sbjct 100 RVITIMQNPRQYSIPDWFLNRQKYVKDGKCSQVLAKGLDNKLPEDLERLKKIRAHRRLHH 159
Query 90 YWGLKVRGQHTKTTGRHGRTVGVAKK 115
+WGL+VRGQHTKTTGR GRT+G++KK
Sbjct 160 FWGLRVRGQHTKTTGRRGRTMGISKK 185
> sce:YML026C RPS18B; Rps18bp; K02964 small subunit ribosomal
protein S18e
Length=146
Score = 116 bits (291), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 50/78 (64%), Positives = 63/78 (80%), Gaps = 0/78 (0%)
Query 28 LLQIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGL 87
L +IV I+ +PT + IP+WFLNRQ D+ DGK H ANN++S R+D ERLKK+R HRG+
Sbjct 66 LERIVQIMQNPTHYKIPAWFLNRQNDITDGKDYHTLANNVESKLRDDLERLKKIRAHRGI 125
Query 88 RHYWGLKVRGQHTKTTGR 105
RH+WGL+VRGQHTKTTGR
Sbjct 126 RHFWGLRVRGQHTKTTGR 143
> sce:YDR450W RPS18A; Rps18ap; K02964 small subunit ribosomal
protein S18e
Length=146
Score = 116 bits (291), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 50/78 (64%), Positives = 63/78 (80%), Gaps = 0/78 (0%)
Query 28 LLQIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGL 87
L +IV I+ +PT + IP+WFLNRQ D+ DGK H ANN++S R+D ERLKK+R HRG+
Sbjct 66 LERIVQIMQNPTHYKIPAWFLNRQNDITDGKDYHTLANNVESKLRDDLERLKKIRAHRGI 125
Query 88 RHYWGLKVRGQHTKTTGR 105
RH+WGL+VRGQHTKTTGR
Sbjct 126 RHFWGLRVRGQHTKTTGR 143
> mmu:384525 Gm5321, EG384525; predicted gene 5321
Length=148
Score = 114 bits (286), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 54/85 (63%), Positives = 65/85 (76%), Gaps = 0/85 (0%)
Query 31 IVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRHY 90
++ I+ +P Q+ IP WFLNRQKDVKDGK R V AN LD+ RED E LKK+R HRGL H+
Sbjct 63 VITIMQNPQQYKIPDWFLNRQKDVKDGKYRQVLANGLDNKLREDLEWLKKIRAHRGLHHF 122
Query 91 WGLKVRGQHTKTTGRHGRTVGVAKK 115
WG +V QHTKTTGR G T+GV+KK
Sbjct 123 WGHRVCCQHTKTTGRRGCTMGVSKK 147
> mmu:629591 Gm6987, EG629591; predicted pseudogene 6987
Length=145
Score = 99.0 bits (245), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 55/111 (49%), Positives = 66/111 (59%), Gaps = 7/111 (6%)
Query 9 SAAHCCCSPAAAALLLLLLLLQIVAIVASPTQFSIPS----WFLNRQKDVKDGKSRHVSA 64
S AH L + ++ I+ +P Q+ IP+ WFLNRQKD GK V
Sbjct 37 SYAHVVLRKTDIGELTEDGVEHLITIMQNPRQYRIPAPPPHWFLNRQKD---GKYSQVLV 93
Query 65 NNLDSVWREDFERLKKMRLHRGLRHYWGLKVRGQHTKTTGRHGRTVGVAKK 115
N LD+ RED ERLKK+R H GL H WGL++ GQHTKTTG GRTVGV KK
Sbjct 94 NGLDNKLREDPERLKKIRAHSGLLHLWGLRIHGQHTKTTGHGGRTVGVFKK 144
> eco:b3298 rpsM, ECK3285, JW3260; 30S ribosomal subunit protein
S13; K02952 small subunit ribosomal protein S13
Length=118
Score = 36.2 bits (82), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 28/52 (53%), Gaps = 0/52 (0%)
Query 54 VKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRHYWGLKVRGQHTKTTGR 105
++D ++ V +L +RL + +RGLRH GL VRGQ TKT R
Sbjct 56 LRDEVAKFVVEGDLRREISMSIKRLMDLGCYRGLRHRRGLPVRGQRTKTNAR 107
> ath:AT1G77750 30S ribosomal protein S13, chloroplast, putative;
K02952 small subunit ribosomal protein S13
Length=154
Score = 33.5 bits (75), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 33/65 (50%), Gaps = 2/65 (3%)
Query 53 DVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRHYWGLKVRGQHTKTTGR--HGRTV 110
D+++ +H + L + +RL ++ +RG RH GL RGQ T T R G+ V
Sbjct 84 DLREEVGQHQHGDELRRRVGSEIQRLVEVDCYRGSRHRHGLPCRGQRTSTNARTKKGKAV 143
Query 111 GVAKK 115
+A K
Sbjct 144 AIAGK 148
> dre:678524 MGC136872; zgc:136872
Length=693
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 24/38 (63%), Gaps = 3/38 (7%)
Query 51 QKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLR 88
+++VKDG+ SANN + +DF++LK+ L LR
Sbjct 13 ERNVKDGQG---SANNPEKFLDQDFQQLKQFCLKERLR 47
> pfa:PF07_0101 conserved Plasmodium protein, unknown function
Length=2190
Score = 28.5 bits (62), Expect = 5.6, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 48 LNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGL 87
LN+ K++ D S NL V++E E KKM + +G
Sbjct 1447 LNKLKNMYDNGSTRKQEINLIRVYKEKVEEFKKMEIIKGF 1486
> sce:YNL081C SWS2; Putative mitochondrial ribosomal protein of
the small subunit, has similarity to E. coli S13 ribosomal
protein; participates in controlling sporulation efficiency;
K02952 small subunit ribosomal protein S13
Length=143
Score = 27.7 bits (60), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 12/40 (30%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 69 SVWREDFERLKKMRLHRGLRHYWGLKVRGQHTKTTGRHGR 108
++ +++ +K+ + G+RH L VRGQHT+ + R
Sbjct 73 AIVKDNIALKRKIGSYSGMRHTLHLPVRGQHTRNNAKTAR 112
> ath:AT4G24900 hypothetical protein
Length=421
Score = 27.7 bits (60), Expect = 8.5, Method: Composition-based stats.
Identities = 11/34 (32%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 45 SWFLNRQKDVKDGKSRHVSANNLDSVWREDFERL 78
+W R+ +++ KS HV+ +N+D W +F R+
Sbjct 339 AWAERRKIEIEMEKSGHVTKSNIDPDWLPNFGRV 372
Lambda K H
0.324 0.133 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2042676840
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40