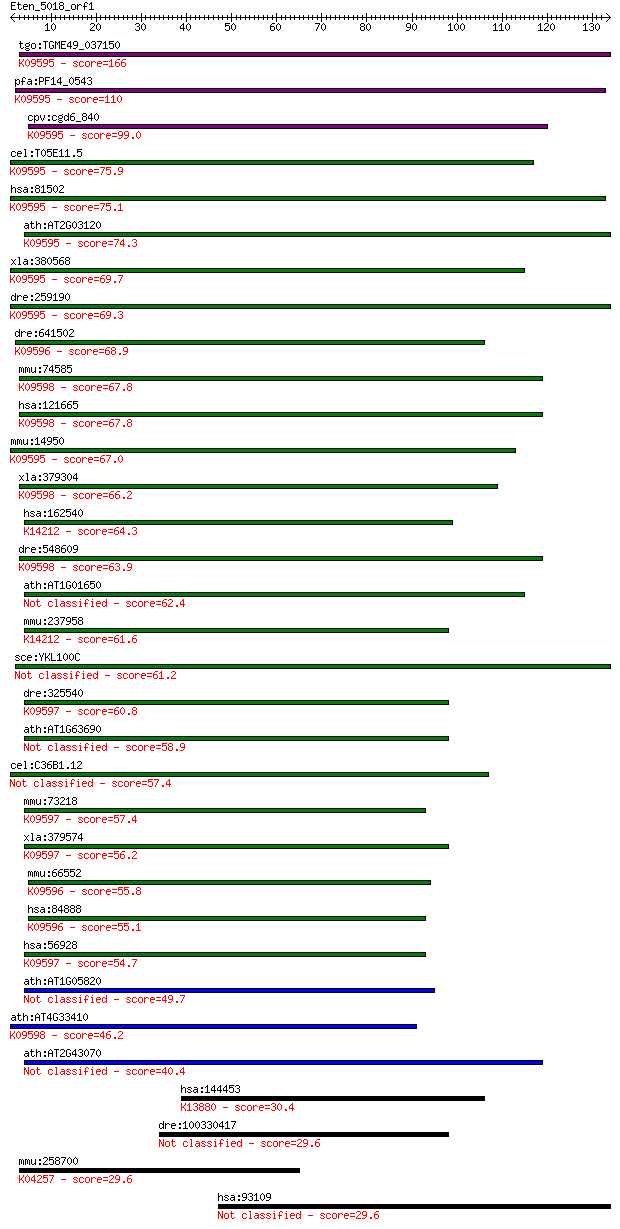
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5018_orf1
Length=133
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_037150 signal peptide peptidase domain-containing p... 166 2e-41
pfa:PF14_0543 SPP; signal peptide peptidase; K09595 minor hist... 110 1e-24
cpv:cgd6_840 shanti/Ykl100cp/Minor histocompatibility antigen ... 99.0 3e-21
cel:T05E11.5 imp-2; IntraMembrane Protease (IMPAS) family memb... 75.9 4e-14
hsa:81502 HM13, H13, IMP1, IMPAS, IMPAS-1, MSTP086, PSENL3, PS... 75.1 5e-14
ath:AT2G03120 ATSPP; ATSPP (ARABIDOPSIS SIGNAL PEPTIDE PEPTIDA... 74.3 8e-14
xla:380568 hm13, MGC68919, h13; histocompatibility (minor) 13 ... 69.7 2e-12
dre:259190 hm13, H13, SPP, cb228, wu:fc11a06, zgc:56660, zgc:8... 69.3 3e-12
dre:641502 MGC123258; zgc:123258; K09596 signal peptide peptid... 68.9 4e-12
mmu:74585 Sppl3, 4833416I09Rik, MGC90867, Psl4, Usmg3; signal ... 67.8 1e-11
hsa:121665 SPPL3, DKFZp586C1324, IMP2, MDHV1887, MGC126674, MG... 67.8 1e-11
mmu:14950 H13, 1200006O09Rik, 4930443L17Rik, 5031424B04Rik, AV... 67.0 1e-11
xla:379304 sppl3, MGC52975; signal peptide peptidase 3; K09598... 66.2 3e-11
hsa:162540 IMP5, SPPL2c; intramembrane protease 5; K14212 sign... 64.3 1e-10
dre:548609 sppl3, fb94d08, wu:fb94d08, zgc:114093; signal pept... 63.9 1e-10
ath:AT1G01650 aspartic-type endopeptidase/ peptidase 62.4 3e-10
mmu:237958 4933407P14Rik, IMP5, Sppl2c; RIKEN cDNA 4933407P14 ... 61.6 6e-10
sce:YKL100C Putative protein of unknown function with similari... 61.2 9e-10
dre:325540 sppl2, SPPL2B, wu:fc16e01, wu:fc85d12, zgc:136525; ... 60.8 1e-09
ath:AT1G63690 protease-associated (PA) domain-containing protein 58.9 4e-09
cel:C36B1.12 imp-1; IntraMembrane Protease (IMPAS) family memb... 57.4 1e-08
mmu:73218 3110056O03Rik, AW550292, PSL1, Sppl2b; RIKEN cDNA 31... 57.4 1e-08
xla:379574 sppl2b, MGC69113; signal peptide peptidase-like 2B;... 56.2 3e-08
mmu:66552 2010106G01Rik, C130089K23Rik, Sppl2A; RIKEN cDNA 201... 55.8 3e-08
hsa:84888 SPPL2A, IMP3, PSL2; signal peptide peptidase-like 2A... 55.1 6e-08
hsa:56928 SPPL2B, IMP4, KIAA1532, MGC111084, PSL1; signal pept... 54.7 8e-08
ath:AT1G05820 aspartic-type endopeptidase/ peptidase 49.7 3e-06
ath:AT4G33410 signal peptide peptidase family protein; K09598 ... 46.2 3e-05
ath:AT2G43070 protease-associated (PA) domain-containing protein 40.4 0.002
hsa:144453 BEST3, MGC13168, MGC40411, VMD2L3; bestrophin 3; K1... 30.4 1.6
dre:100330417 5-AMP-activated protein kinase subunit gamma-1-like 29.6 2.3
mmu:258700 Olfr1451, MOR202-1; olfactory receptor 1451; K04257... 29.6 2.4
hsa:93109 TMEM44, DKFZp686O18124, MGC131692, MGC163169; transm... 29.6 2.9
> tgo:TGME49_037150 signal peptide peptidase domain-containing
protein ; K09595 minor histocompatibility antigen H13 [EC:3.4.23.-]
Length=417
Score = 166 bits (419), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 84/137 (61%), Positives = 101/137 (73%), Gaps = 6/137 (4%)
Query 3 QYSILGLGDVVIPGVLIAMCLRFDLFLYQ---KQQNAA---TKALAVDIHQRFPKFYFCV 56
Q+SILGLGD+VIPGV I+MCLRFD L NAA T ++DIHQ+F KFYF V
Sbjct 281 QHSILGLGDIVIPGVFISMCLRFDYSLATASVTNGNAAKTTTVGASIDIHQKFSKFYFFV 340
Query 57 VLAFYELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEEDEEG 116
V FYE GLLTTG++ML QHPQPALLY+VP+CLFSLFGAAALNG+VKEVLAY+E++EE
Sbjct 341 VSIFYEFGLLTTGVIMLVFQHPQPALLYIVPFCLFSLFGAAALNGQVKEVLAYREDEEEK 400
Query 117 SSILRASALETTEKKEN 133
+ + + EKK
Sbjct 401 PAEVEGESEMKEEKKRK 417
> pfa:PF14_0543 SPP; signal peptide peptidase; K09595 minor histocompatibility
antigen H13 [EC:3.4.23.-]
Length=412
Score = 110 bits (274), Expect = 1e-24, Method: Composition-based stats.
Identities = 51/134 (38%), Positives = 88/134 (65%), Gaps = 3/134 (2%)
Query 2 LQYSILGLGDVVIPGVLIAMCLRFDLFLYQK---QQNAATKALAVDIHQRFPKFYFCVVL 58
+ YS+LGLGD++IPG+L+++CLRFD +L++ + N + IH+ F K+YF ++
Sbjct 260 VHYSMLGLGDIIIPGILMSLCLRFDYYLFKNNIHKGNLKKMFNDISIHESFKKYYFYTII 319
Query 59 AFYELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEEDEEGSS 118
FYELGL+ T ++ Y +HPQPALLYLVP C+ ++ + + K ++ Y+E ++ ++
Sbjct 320 IFYELGLVVTYCMLFYFEHPQPALLYLVPACILAILACSICKREFKLMIKYQEITDKSNT 379
Query 119 ILRASALETTEKKE 132
+ AS + +K+E
Sbjct 380 VDDASKNKKKDKEE 393
> cpv:cgd6_840 shanti/Ykl100cp/Minor histocompatibility antigen
H13-like; presenilin, signal peptide peptidase family, with
10 transmembrane domains and a signal peptide ; K09595 minor
histocompatibility antigen H13 [EC:3.4.23.-]
Length=408
Score = 99.0 bits (245), Expect = 3e-21, Method: Composition-based stats.
Identities = 54/115 (46%), Positives = 67/115 (58%), Gaps = 3/115 (2%)
Query 5 SILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYELG 64
SILGLGD+VIPG+ I++CLRFDL Y K+ N + L I FC VL Y LG
Sbjct 266 SILGLGDIVIPGLFISLCLRFDLKDYTKKHNQSLYHL---ISSSLQTPTFCTVLVSYLLG 322
Query 65 LLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEEDEEGSSI 119
L+TT VMLY + QPALLYLVP+CL S+ + K + Y EE + I
Sbjct 323 LITTACVMLYFKAAQPALLYLVPFCLISMVLSVVYRNKSSDAWNYSEEADSDERI 377
> cel:T05E11.5 imp-2; IntraMembrane Protease (IMPAS) family member
(imp-2); K09595 minor histocompatibility antigen H13 [EC:3.4.23.-]
Length=468
Score = 75.9 bits (185), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 48/120 (40%), Positives = 67/120 (55%), Gaps = 9/120 (7%)
Query 1 ALQYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAF 60
A ++S+LGLGD+VIPG+ IA+ RFD + Q A +KA + R+ YF V +
Sbjct 343 ASKHSMLGLGDIVIPGIFIALLRRFDYRVVQ--TTAESKAPQGSLKGRY---YFVVTVVA 397
Query 61 YELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKE----EDEEG 116
Y GL T VM + + QPALLYLVP CLF A + G++ + Y E ++EE
Sbjct 398 YMAGLFITMAVMHHFKAAQPALLYLVPCCLFVPLLLAVIRGELSALWNYDESRHVDNEEN 457
> hsa:81502 HM13, H13, IMP1, IMPAS, IMPAS-1, MSTP086, PSENL3,
PSL3, SPP, dJ324O17.1; histocompatibility (minor) 13; K09595
minor histocompatibility antigen H13 [EC:3.4.23.-]
Length=377
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 55/142 (38%), Positives = 73/142 (51%), Gaps = 30/142 (21%)
Query 1 ALQYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAF 60
A +++LGLGDVVIPG+ IA+ LRFD+ L ++N T YF A
Sbjct 255 ANNFAMLGLGDVVIPGIFIALLLRFDISL---KKNTHT--------------YFYTSFAA 297
Query 61 YELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKE--------- 111
Y GL T +M +H QPALLYLVP C+ A G+V E+ +Y+E
Sbjct 298 YIFGLGLTIFIMHIFKHAQPALLYLVPACIGFPVLVALAKGEVTEMFSYEESNPKDPAAV 357
Query 112 -EDEEGSSILRASALETTEKKE 132
E +EG+ ASA + EKKE
Sbjct 358 TESKEGT---EASASKGLEKKE 376
> ath:AT2G03120 ATSPP; ATSPP (ARABIDOPSIS SIGNAL PEPTIDE PEPTIDASE);
aspartic-type endopeptidase; K09595 minor histocompatibility
antigen H13 [EC:3.4.23.-]
Length=344
Score = 74.3 bits (181), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 43/130 (33%), Positives = 65/130 (50%), Gaps = 18/130 (13%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYEL 63
YS+LGLGD+VIPG+ +A+ LRFD+ ++ Q YF Y +
Sbjct 232 YSMLGLGDIVIPGIFVALALRFDVSRRRQPQ------------------YFTSAFIGYAV 273
Query 64 GLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEEDEEGSSILRAS 123
G++ T +VM + Q QPALLY+VP + L NG +K +LA+ E E ++ +
Sbjct 274 GVILTIVVMNWFQAAQPALLYIVPAVIGFLASHCIWNGDIKPLLAFDESKTEEATTDESK 333
Query 124 ALETTEKKEN 133
E K +
Sbjct 334 TSEEVNKAHD 343
> xla:380568 hm13, MGC68919, h13; histocompatibility (minor) 13
(EC:3.4.99.-); K09595 minor histocompatibility antigen H13
[EC:3.4.23.-]
Length=392
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 44/114 (38%), Positives = 61/114 (53%), Gaps = 17/114 (14%)
Query 1 ALQYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAF 60
A +++LGLGD+VIPG+ IA+ LRFD+ L ++N+ T YF
Sbjct 253 ANNFAMLGLGDIVIPGIFIALLLRFDVSL---KKNSHT--------------YFYTSFLA 295
Query 61 YELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEEDE 114
Y GL T VM +H QPALLYLVP C+ A + G+V E+ +Y+ E
Sbjct 296 YVFGLALTIFVMHTFKHAQPALLYLVPACIGFPLLVALVKGEVTEMFSYESSAE 349
> dre:259190 hm13, H13, SPP, cb228, wu:fc11a06, zgc:56660, zgc:86862;
histocompatibility (minor) 13 (EC:3.4.99.-); K09595
minor histocompatibility antigen H13 [EC:3.4.23.-]
Length=366
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 49/133 (36%), Positives = 68/133 (51%), Gaps = 25/133 (18%)
Query 1 ALQYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAF 60
A +++LGLGD+VIPG+ IA+ LRFD+ L ++N T YF
Sbjct 258 ASNFAMLGLGDIVIPGIFIALLLRFDVSL---KKNTRT--------------YFYTSFLA 300
Query 61 YELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEEDEEGSSIL 120
Y GL T VM +H QPALLYLVP C+ A + G++ E+ Y+EE
Sbjct 301 YIFGLGLTIFVMHTFKHAQPALLYLVPACVGFPVLVALVKGELTEMFRYEEE-------- 352
Query 121 RASALETTEKKEN 133
S ETTE +++
Sbjct 353 TPSKEETTESEKD 365
> dre:641502 MGC123258; zgc:123258; K09596 signal peptide peptidase-like
2A [EC:3.4.23.-]
Length=519
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 40/104 (38%), Positives = 57/104 (54%), Gaps = 17/104 (16%)
Query 2 LQYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFY 61
+Q+SILG GD++IPG+L+A C RFD+++ + K YF Y
Sbjct 416 MQFSILGYGDIIIPGLLVAYCHRFDVWVGNSR-----------------KTYFITCAVAY 458
Query 62 ELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKE 105
+GLL T VML ++ QPALLYLVP L S F A + +++
Sbjct 459 AVGLLLTFAVMLLSRMGQPALLYLVPCTLLSSFTLACVRKELRH 502
> mmu:74585 Sppl3, 4833416I09Rik, MGC90867, Psl4, Usmg3; signal
peptide peptidase 3; K09598 signal peptide peptidase-like
3 [EC:3.4.23.-]
Length=384
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 40/117 (34%), Positives = 61/117 (52%), Gaps = 1/117 (0%)
Query 3 QYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKF-YFCVVLAFY 61
+S+LG+GD+V+PG+L+ LR+D + Q ++ +I R K YF L Y
Sbjct 263 HFSMLGIGDIVMPGLLLCFVLRYDNYKKQASGDSCGAPGPANISGRMQKVSYFHCTLIGY 322
Query 62 ELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEEDEEGSS 118
+GLLT + + QPALLYLVP+ L L A L G ++ + + + SS
Sbjct 323 FVGLLTATVASRIHRAAQPALLYLVPFTLLPLLTMAYLKGDLRRMWSEPFHSKSSSS 379
> hsa:121665 SPPL3, DKFZp586C1324, IMP2, MDHV1887, MGC126674,
MGC126676, MGC90402, PRO4332, PSL4; signal peptide peptidase
3; K09598 signal peptide peptidase-like 3 [EC:3.4.23.-]
Length=384
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 40/117 (34%), Positives = 61/117 (52%), Gaps = 1/117 (0%)
Query 3 QYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKF-YFCVVLAFY 61
+S+LG+GD+V+PG+L+ LR+D + Q ++ +I R K YF L Y
Sbjct 263 HFSMLGIGDIVMPGLLLCFVLRYDNYKKQASGDSCGAPGPANISGRMQKVSYFHCTLIGY 322
Query 62 ELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEEDEEGSS 118
+GLLT + + QPALLYLVP+ L L A L G ++ + + + SS
Sbjct 323 FVGLLTATVASRIHRAAQPALLYLVPFTLLPLLTMAYLKGDLRRMWSEPFHSKSSSS 379
> mmu:14950 H13, 1200006O09Rik, 4930443L17Rik, 5031424B04Rik,
AV020344, H-13, Hm13, PSL3, Spp; histocompatibility 13 (EC:3.4.99.-);
K09595 minor histocompatibility antigen H13 [EC:3.4.23.-]
Length=394
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 43/112 (38%), Positives = 59/112 (52%), Gaps = 17/112 (15%)
Query 1 ALQYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAF 60
A +++LGLGD+VIPG+ IA+ LRFD+ L ++N T YF A
Sbjct 255 ADNFAMLGLGDIVIPGIFIALLLRFDISL---KKNTHT--------------YFYTSFAA 297
Query 61 YELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEE 112
Y GL T +M +H QPALLYLVP C+ A G+V E+ +Y+
Sbjct 298 YIFGLGLTIFIMHIFKHAQPALLYLVPACIGFPVLVALAKGEVAEMFSYESS 349
> xla:379304 sppl3, MGC52975; signal peptide peptidase 3; K09598
signal peptide peptidase-like 3 [EC:3.4.23.-]
Length=379
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 62/106 (58%), Gaps = 3/106 (2%)
Query 3 QYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYE 62
+S+LG+GD+V+PG+L+ LR+D Y+KQ + ++ + + ++ C ++ ++
Sbjct 262 HFSMLGIGDIVMPGLLLCFVLRYDN--YKKQATSDSQGAPISGRMQKVSYFHCTLIGYF- 318
Query 63 LGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLA 108
+GLLT + + QPALLYLVP+ L L A L G ++ + +
Sbjct 319 VGLLTATVASRIHRAAQPALLYLVPFTLLPLLTMAYLKGDLRRMWS 364
> hsa:162540 IMP5, SPPL2c; intramembrane protease 5; K14212 signal
peptide peptidase-like 2C [EC:3.4.23.-]
Length=684
Score = 64.3 bits (155), Expect = 1e-10, Method: Composition-based stats.
Identities = 36/95 (37%), Positives = 47/95 (49%), Gaps = 18/95 (18%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYEL 63
+SILG GD+V+PG L+A C RFD+ + +Q YF Y +
Sbjct 441 FSILGFGDIVVPGFLVAYCCRFDVQVCSRQ------------------IYFVACTVAYAV 482
Query 64 GLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAA 98
GLL T + M+ Q QPALLYLV L + AA
Sbjct 483 GLLVTFMAMVLMQMGQPALLYLVSSTLLTSLAVAA 517
> dre:548609 sppl3, fb94d08, wu:fb94d08, zgc:114093; signal peptide
peptidase 3; K09598 signal peptide peptidase-like 3 [EC:3.4.23.-]
Length=382
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 41/117 (35%), Positives = 62/117 (52%), Gaps = 4/117 (3%)
Query 3 QYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKF-YFCVVLAFY 61
+S+LG+GD+V+PG+L+ LR+D Y+KQ A ++ R + YF L Y
Sbjct 264 HFSMLGIGDIVMPGLLLCFVLRYDN--YKKQATGEVPGPA-NMSGRMQRVSYFHCTLIGY 320
Query 62 ELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEEDEEGSS 118
+GLLT + + QPALLYLVP+ L L A L G ++ + + + SS
Sbjct 321 FVGLLTATVASRIHRAAQPALLYLVPFTLLPLLTMAYLKGDLRRMWSEPFHAKSSSS 377
> ath:AT1G01650 aspartic-type endopeptidase/ peptidase
Length=398
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 38/112 (33%), Positives = 56/112 (50%), Gaps = 17/112 (15%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYEL 63
YSI+G GD+++PG+L+ LR+D ++R YF ++ Y L
Sbjct 291 YSIIGFGDIILPGLLVTFALRYDWL----------------ANKRLKSGYFLGTMSAYGL 334
Query 64 GLLTTGLVM-LYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEEDE 114
GLL T + + L H QPALLY+VP+ L +LF G +K + E D
Sbjct 335 GLLITYIALNLMDGHGQPALLYIVPFILGTLFVLGHKRGDLKTLWTTGEPDR 386
> mmu:237958 4933407P14Rik, IMP5, Sppl2c; RIKEN cDNA 4933407P14
gene; K14212 signal peptide peptidase-like 2C [EC:3.4.23.-]
Length=581
Score = 61.6 bits (148), Expect = 6e-10, Method: Composition-based stats.
Identities = 34/94 (36%), Positives = 46/94 (48%), Gaps = 18/94 (19%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYEL 63
+SILG GD+V+PG L+A C RFD+ + +Q Y+ Y +
Sbjct 450 FSILGFGDIVVPGFLVAYCHRFDMQVQSRQ------------------VYYMACTVAYAV 491
Query 64 GLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAA 97
GLL T + M+ Q QPALLYLV L + A
Sbjct 492 GLLVTFVAMILMQMGQPALLYLVSSTLLTSLAVA 525
> sce:YKL100C Putative protein of unknown function with similarity
to a human minor histocompatibility antigen and signal
peptide peptidases; YKL100C is not an essential gene
Length=587
Score = 61.2 bits (147), Expect = 9e-10, Method: Composition-based stats.
Identities = 38/132 (28%), Positives = 63/132 (47%), Gaps = 7/132 (5%)
Query 2 LQYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFY 61
+SILGLGD+ +PG+ IAMC ++D++ + + ++ + YF + Y
Sbjct 402 FNFSILGLGDIALPGMFIAMCYKYDIWKWHLDHDDTEFHF---LNWSYVGKYFITAMVSY 458
Query 62 ELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEEDEEGSSILR 121
L++ + + QPALLY+VP L S A N K+ ++ + E L+
Sbjct 459 VASLVSAMVSLSIFNTAQPALLYIVPSLLISTILVACWNKDFKQFWNFQYDTIEVDKSLK 518
Query 122 ASALETTEKKEN 133
+ EKKEN
Sbjct 519 ----KAIEKKEN 526
> dre:325540 sppl2, SPPL2B, wu:fc16e01, wu:fc85d12, zgc:136525;
signal peptide peptidase-like 2; K09597 signal peptide peptidase-like
2B [EC:3.4.23.-]
Length=564
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 33/94 (35%), Positives = 46/94 (48%), Gaps = 18/94 (19%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYEL 63
+S+LG GD+++PG+L+A C RFD+ + Q YF Y +
Sbjct 416 FSLLGFGDILVPGLLVAYCHRFDILMQTSQ------------------IYFLACTIGYGI 457
Query 64 GLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAA 97
GLL T + + Q QPALLYLVP L + A
Sbjct 458 GLLITFVALTLMQMGQPALLYLVPCTLLTSLAVA 491
> ath:AT1G63690 protease-associated (PA) domain-containing protein
Length=540
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 37/95 (38%), Positives = 50/95 (52%), Gaps = 17/95 (17%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYEL 63
YSI+G GD+++PG+LIA LR+D A K L YF + Y L
Sbjct 431 YSIIGFGDILLPGLLIAFALRYDWL--------ANKTLRTG--------YFIWAMVAYGL 474
Query 64 GLLTTGLVM-LYAQHPQPALLYLVPYCLFSLFGAA 97
GLL T + + L H QPALLY+VP+ L ++ A
Sbjct 475 GLLITYVALNLMDGHGQPALLYIVPFTLGTMLTLA 509
> cel:C36B1.12 imp-1; IntraMembrane Protease (IMPAS) family member
(imp-1)
Length=662
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 33/106 (31%), Positives = 51/106 (48%), Gaps = 17/106 (16%)
Query 1 ALQYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAF 60
Q++ILGLGD+V+PG L+A C + F +R Y + +
Sbjct 506 GFQFTILGLGDIVMPGYLVAHCFTMNGF-----------------SERVRLIYGFISVVG 548
Query 61 YELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEV 106
Y +GL+ T L + + QPAL+YLVP LF + A G+ ++
Sbjct 549 YGIGLIVTFLALALMKTAQPALIYLVPSTLFPIIMLALCRGEFLKI 594
> mmu:73218 3110056O03Rik, AW550292, PSL1, Sppl2b; RIKEN cDNA
3110056O03 gene (EC:3.4.23.-); K09597 signal peptide peptidase-like
2B [EC:3.4.23.-]
Length=578
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 34/89 (38%), Positives = 48/89 (53%), Gaps = 18/89 (20%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYEL 63
+S+LG GD+++PG+L+A C RFD+ Q Q + + YF Y L
Sbjct 407 FSLLGFGDILVPGLLVAYCHRFDI---QVQSS---------------RIYFVACTIAYGL 448
Query 64 GLLTTGLVMLYAQHPQPALLYLVPYCLFS 92
GLL T + ++ Q QPALLYLVP L +
Sbjct 449 GLLVTFVALVLMQRGQPALLYLVPCTLLT 477
> xla:379574 sppl2b, MGC69113; signal peptide peptidase-like 2B;
K09597 signal peptide peptidase-like 2B [EC:3.4.23.-]
Length=606
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 34/94 (36%), Positives = 48/94 (51%), Gaps = 18/94 (19%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYEL 63
+S+LG GD+++PG+L+A C RFD+ Q Q + + YF Y +
Sbjct 387 FSLLGFGDILVPGLLVAYCHRFDI---QVQSS---------------RIYFVACTIAYGI 428
Query 64 GLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAA 97
GLL T + + Q QPALLYLVP L + A
Sbjct 429 GLLLTFVALALMQKGQPALLYLVPCTLLTCLAVA 462
> mmu:66552 2010106G01Rik, C130089K23Rik, Sppl2A; RIKEN cDNA 2010106G01
gene (EC:3.4.23.-); K09596 signal peptide peptidase-like
2A [EC:3.4.23.-]
Length=523
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 30/89 (33%), Positives = 46/89 (51%), Gaps = 19/89 (21%)
Query 5 SILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYELG 64
S+LG GD+++PG+LIA C RFD+ Q Y+ Y +G
Sbjct 410 SVLGFGDIIVPGLLIAYCRRFDV-------------------QTGSSIYYISSTIAYAVG 450
Query 65 LLTTGLVMLYAQHPQPALLYLVPYCLFSL 93
++ T +V++ + QPALLYLVP L ++
Sbjct 451 MIITFVVLMVMKTGQPALLYLVPCTLITV 479
> hsa:84888 SPPL2A, IMP3, PSL2; signal peptide peptidase-like
2A (EC:3.4.23.-); K09596 signal peptide peptidase-like 2A [EC:3.4.23.-]
Length=520
Score = 55.1 bits (131), Expect = 6e-08, Method: Composition-based stats.
Identities = 31/88 (35%), Positives = 47/88 (53%), Gaps = 18/88 (20%)
Query 5 SILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYELG 64
SILG GD+++PG+LIA C RFD+ Q ++ Y+ Y +G
Sbjct 406 SILGFGDIIVPGLLIAYCRRFDV------QTGSS------------YIYYVSSTVAYAIG 447
Query 65 LLTTGLVMLYAQHPQPALLYLVPYCLFS 92
++ T +V++ + QPALLYLVP L +
Sbjct 448 MILTFVVLVLMKKGQPALLYLVPCTLIT 475
> hsa:56928 SPPL2B, IMP4, KIAA1532, MGC111084, PSL1; signal peptide
peptidase-like 2B (EC:3.4.23.-); K09597 signal peptide
peptidase-like 2B [EC:3.4.23.-]
Length=511
Score = 54.7 bits (130), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 33/89 (37%), Positives = 47/89 (52%), Gaps = 18/89 (20%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYEL 63
+S+LG GD+++PG+L+A C RFD+ Q Q + + YF Y +
Sbjct 414 FSLLGFGDILVPGLLVAYCHRFDI---QVQSS---------------RVYFVACTIAYGV 455
Query 64 GLLTTGLVMLYAQHPQPALLYLVPYCLFS 92
GLL T + + Q QPALLYLVP L +
Sbjct 456 GLLVTFVALALMQRGQPALLYLVPCTLVT 484
> ath:AT1G05820 aspartic-type endopeptidase/ peptidase
Length=507
Score = 49.7 bits (117), Expect = 3e-06, Method: Composition-based stats.
Identities = 34/92 (36%), Positives = 45/92 (48%), Gaps = 17/92 (18%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYEL 63
Y+++G GD++ PG+LI RFD K+ N YF ++ Y L
Sbjct 430 YNMIGFGDILFPGLLICFIFRFD-----KENNKGVS-----------NGYFPWLMFGYGL 473
Query 64 GLLTTGL-VMLYAQHPQPALLYLVPYCLFSLF 94
GL T L + + H QPALLYLVP L LF
Sbjct 474 GLFLTYLGLYVMNGHGQPALLYLVPCTLGILF 505
> ath:AT4G33410 signal peptide peptidase family protein; K09598
signal peptide peptidase-like 3 [EC:3.4.23.-]
Length=372
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 45/90 (50%), Gaps = 6/90 (6%)
Query 1 ALQYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAF 60
A + +LGLGD+ IP +L+A+ L FD ++K ++ D+ Y L
Sbjct 255 ASDFMMLGLGDMAIPAMLLALVLCFD---HRKTRDVVN---IFDLKSSKGHKYIWYALPG 308
Query 61 YELGLLTTGLVMLYAQHPQPALLYLVPYCL 90
Y +GL+ + PQPALLYLVP L
Sbjct 309 YAIGLVAALAAGVLTHSPQPALLYLVPSTL 338
> ath:AT2G43070 protease-associated (PA) domain-containing protein
Length=540
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 31/115 (26%), Positives = 52/115 (45%), Gaps = 15/115 (13%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYEL 63
Y ++G GD++ PG+LI+ R+D + N ++ + + +
Sbjct 433 YDMIGFGDILFPGLLISFASRYDKIKKRVISNG---------------YFLWLTIGYGIG 477
Query 64 GLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEEDEEGSS 118
LLT + L H QPALLY+VP L + G++KE+ Y E+ E +
Sbjct 478 LLLTYLGLYLMDGHGQPALLYIVPCTLGLAVILGLVRGELKELWNYGIEESESHT 532
> hsa:144453 BEST3, MGC13168, MGC40411, VMD2L3; bestrophin 3;
K13880 bestrophin-3
Length=668
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 18/68 (26%), Positives = 32/68 (47%), Gaps = 6/68 (8%)
Query 39 KALAVDIHQRFPKFYFCVVLAFYELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAA 98
++++ +++RFP V E G +TT L+ P L Y VP+ F A
Sbjct 141 RSVSTAVYKRFPTMDHVV-----EAGFMTTDERKLFNHLKSPHLKYWVPFIWFGNLATKA 195
Query 99 LN-GKVKE 105
N G++++
Sbjct 196 RNEGRIRD 203
> dre:100330417 5-AMP-activated protein kinase subunit gamma-1-like
Length=211
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 33/66 (50%), Gaps = 2/66 (3%)
Query 34 QNAATKALAVDIHQRFPKFYFC--VVLAFYELGLLTTGLVMLYAQHPQPALLYLVPYCLF 91
++ A + DI+ RF K + C +V +L + T L H + ++PYC+F
Sbjct 114 EDEAVEHSESDIYMRFMKSHKCYDIVPTSSKLVVFDTTLQWFAVPHIDFFVSAVLPYCVF 173
Query 92 SLFGAA 97
S +G++
Sbjct 174 STYGSS 179
> mmu:258700 Olfr1451, MOR202-1; olfactory receptor 1451; K04257
olfactory receptor
Length=310
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 34/64 (53%), Gaps = 4/64 (6%)
Query 3 QYSILGLGDVVIPGVLIAMCLRFDL-FLYQKQQNAATKALA-VDIHQRFPKFYFCVVLAF 60
++ +LGL D PG+ +++C+ F L +L N L +D H P ++F L+F
Sbjct 9 EFILLGLTD--DPGLQVSLCIMFTLIYLIDVVGNTGLIMLVLMDSHLHTPMYFFLCNLSF 66
Query 61 YELG 64
+LG
Sbjct 67 VDLG 70
> hsa:93109 TMEM44, DKFZp686O18124, MGC131692, MGC163169; transmembrane
protein 44
Length=428
Score = 29.6 bits (65), Expect = 2.9, Method: Composition-based stats.
Identities = 28/100 (28%), Positives = 44/100 (44%), Gaps = 15/100 (15%)
Query 47 QRFPKFYFCVVLAFYELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALN------ 100
+ FP + L GLL ++ + QHP+ LL P+ L SL G AAL+
Sbjct 208 KTFPSIHLWTRLLSALAGLLYASAIVAHDQHPE-YLLRATPWFLTSL-GRAALDLAIIFL 265
Query 101 -----GKVKEVLAYKEEDEEG--SSILRASALETTEKKEN 133
K+++ L + +E E + L A + E +EN
Sbjct 266 SCVMKSKMRQALGFAKEARESPDTQALLTCAEKEEENQEN 305
Lambda K H
0.325 0.140 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2165519004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40