bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5022_orf1
Length=128
Score E
Sequences producing significant alignments: (Bits) Value
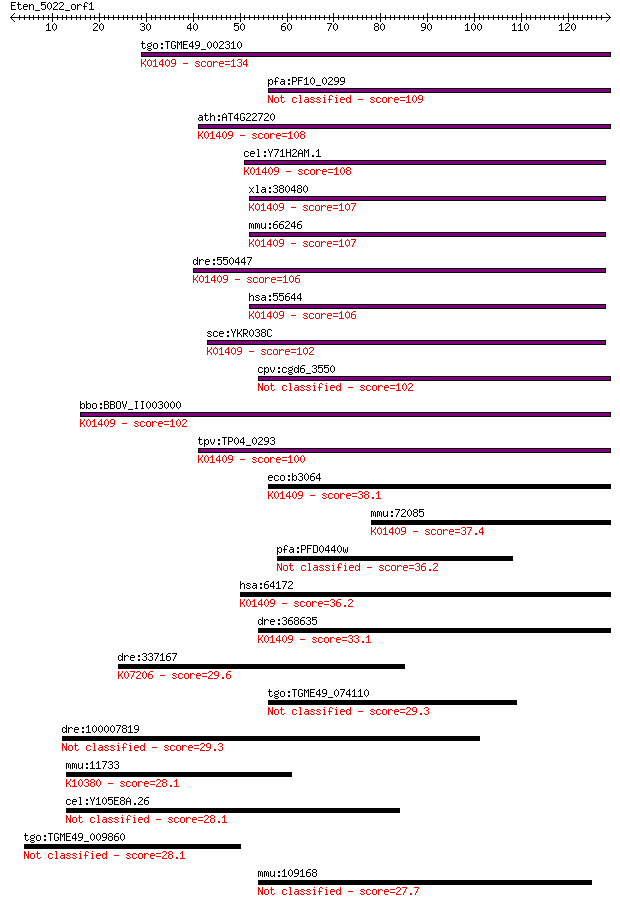
tgo:TGME49_002310 glycoprotease family domain-containing prote... 134 8e-32
pfa:PF10_0299 glycoprotease, putative 109 2e-24
ath:AT4G22720 glycoprotease M22 family protein; K01409 O-sialo... 108 3e-24
cel:Y71H2AM.1 hypothetical protein; K01409 O-sialoglycoprotein... 108 5e-24
xla:380480 osgep, MGC64557; O-sialoglycoprotein endopeptidase ... 107 9e-24
mmu:66246 Osgep, 1500019L24Rik, GCPL-1, PRSMG1; O-sialoglycopr... 107 1e-23
dre:550447 osgep, si:ch211-214j24.11, wu:fj38e02, zgc:112527; ... 106 2e-23
hsa:55644 OSGEP, FLJ20411, GCPL1, KAE1, OSGEP1, PRSMG1; O-sial... 106 2e-23
sce:YKR038C KAE1; Highly conserved putative glycoprotease prop... 102 3e-22
cpv:cgd6_3550 endopeptidase 102 4e-22
bbo:BBOV_II003000 18.m06249; glycoprotease family protein; K01... 102 4e-22
tpv:TP04_0293 hypothetical protein; K01409 O-sialoglycoprotein... 100 1e-21
eco:b3064 gcp, ECK3054, JW3036, ygjD; glycation-binding protei... 38.1 0.007
mmu:72085 Osgepl1, 2610001M19Rik, AA416452, MGC13061; O-sialog... 37.4 0.013
pfa:PFD0440w peptidase, M22 family, putative 36.2 0.024
hsa:64172 OSGEPL1; O-sialoglycoprotein endopeptidase-like 1 (E... 36.2 0.026
dre:368635 osgepl1, MGC123084, si:dz72b14.6; O-sialoglycoprote... 33.1 0.21
dre:337167 tsc1a, fa99f04, wu:fa99f04, zgc:63657; tuberous scl... 29.6 2.3
tgo:TGME49_074110 hypothetical protein 29.3 3.3
dre:100007819 solute carrier family 22 member 6-like 29.3 3.4
mmu:11733 Ank1, Ank-1, nb, pale; ankyrin 1, erythroid; K10380 ... 28.1 6.9
cel:Y105E8A.26 zoo-1; ZO-1 (Zonula Occludens tight junctional ... 28.1 7.2
tgo:TGME49_009860 nucleolar phosphoprotein p130, putative 28.1 7.5
mmu:109168 Atl3, 4633402C03Rik, 5730596K20Rik, AI465397, AW228... 27.7 9.1
> tgo:TGME49_002310 glycoprotease family domain-containing protein
(EC:3.4.24.57); K01409 O-sialoglycoprotein endopeptidase
[EC:3.4.24.57]
Length=580
Score = 134 bits (337), Expect = 8e-32, Method: Compositional matrix adjust.
Identities = 66/104 (63%), Positives = 80/104 (76%), Gaps = 4/104 (3%)
Query 29 GQLRKRGNPSRE--SAD--SSIPEELLTPESICYSLQEYLFGMLVEITERAMALSHSSDV 84
G +GN R+ S D +P LLTPES+C+S QE +F ML E+TERAMAL ++ V
Sbjct 431 GSCAAKGNSKRQCQSVDMFEGLPTCLLTPESLCFSAQEIIFAMLTEVTERAMALHYADQV 490
Query 85 LVVGGVGCNLRLQQMLREMARQRGCSMGGMDERYCIDNGAMIAY 128
LVVGGVGCNLRLQ+ML+EMA +RG SMGGMD+RYCIDNGAM+AY
Sbjct 491 LVVGGVGCNLRLQEMLKEMAMRRGASMGGMDDRYCIDNGAMVAY 534
> pfa:PF10_0299 glycoprotease, putative
Length=598
Score = 109 bits (272), Expect = 2e-24, Method: Composition-based stats.
Identities = 45/73 (61%), Positives = 61/73 (83%), Gaps = 0/73 (0%)
Query 56 ICYSLQEYLFGMLVEITERAMALSHSSDVLVVGGVGCNLRLQQMLREMARQRGCSMGGMD 115
ICYSLQ ++F ML+EITERA+A ++S +V++VGGVGCNL LQ M+++MA+Q+ +G MD
Sbjct 490 ICYSLQHHIFSMLIEITERAIAFTNSKEVIIVGGVGCNLFLQNMMKKMAKQKNIKIGFMD 549
Query 116 ERYCIDNGAMIAY 128
YC+DNGAMIAY
Sbjct 550 HSYCVDNGAMIAY 562
> ath:AT4G22720 glycoprotease M22 family protein; K01409 O-sialoglycoprotein
endopeptidase [EC:3.4.24.57]
Length=353
Score = 108 bits (271), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 51/88 (57%), Positives = 62/88 (70%), Gaps = 0/88 (0%)
Query 41 SADSSIPEELLTPESICYSLQEYLFGMLVEITERAMALSHSSDVLVVGGVGCNLRLQQML 100
+A+ + TP +CYSLQE +F MLVEITERAMA DVL+VGGVGCN RLQ+M+
Sbjct 218 TAEEKLKNNECTPADLCYSLQETVFAMLVEITERAMAHCDKKDVLIVGGVGCNERLQEMM 277
Query 101 REMARQRGCSMGGMDERYCIDNGAMIAY 128
R M +R + D+RYCIDNGAMIAY
Sbjct 278 RTMCSERDGKLFATDDRYCIDNGAMIAY 305
> cel:Y71H2AM.1 hypothetical protein; K01409 O-sialoglycoprotein
endopeptidase [EC:3.4.24.57]
Length=337
Score = 108 bits (269), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 48/77 (62%), Positives = 60/77 (77%), Gaps = 0/77 (0%)
Query 51 LTPESICYSLQEYLFGMLVEITERAMALSHSSDVLVVGGVGCNLRLQQMLREMARQRGCS 110
TPE +C+SLQE +F ML+EITERAMA + S ++L+VGGVGCNLRLQ+M M +RG
Sbjct 226 FTPEDLCFSLQETVFAMLIEITERAMAHTSSKELLIVGGVGCNLRLQEMASAMCAERGAH 285
Query 111 MGGMDERYCIDNGAMIA 127
+ DER+CIDNGAMIA
Sbjct 286 LFATDERFCIDNGAMIA 302
> xla:380480 osgep, MGC64557; O-sialoglycoprotein endopeptidase
(EC:3.4.24.57); K01409 O-sialoglycoprotein endopeptidase [EC:3.4.24.57]
Length=335
Score = 107 bits (267), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 50/76 (65%), Positives = 60/76 (78%), Gaps = 0/76 (0%)
Query 52 TPESICYSLQEYLFGMLVEITERAMALSHSSDVLVVGGVGCNLRLQQMLREMARQRGCSM 111
TPE +C+SLQE LF MLVEITERAMA S +VL+VGGVGCN+RLQ+M+ M +RG +
Sbjct 225 TPEDLCFSLQETLFSMLVEITERAMAHCGSQEVLIVGGVGCNVRLQEMMGVMCEERGAKI 284
Query 112 GGMDERYCIDNGAMIA 127
DER+CIDNGAMIA
Sbjct 285 FATDERFCIDNGAMIA 300
> mmu:66246 Osgep, 1500019L24Rik, GCPL-1, PRSMG1; O-sialoglycoprotein
endopeptidase (EC:3.4.24.57); K01409 O-sialoglycoprotein
endopeptidase [EC:3.4.24.57]
Length=335
Score = 107 bits (266), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 48/76 (63%), Positives = 60/76 (78%), Gaps = 0/76 (0%)
Query 52 TPESICYSLQEYLFGMLVEITERAMALSHSSDVLVVGGVGCNLRLQQMLREMARQRGCSM 111
TPE +C+SLQE +F MLVEITERAMA S + L+VGGVGCNLRLQ+M+ M ++RG +
Sbjct 225 TPEDLCFSLQETVFAMLVEITERAMAHCGSKEALIVGGVGCNLRLQEMMGTMCQERGAQL 284
Query 112 GGMDERYCIDNGAMIA 127
DER+C+DNGAMIA
Sbjct 285 FATDERFCVDNGAMIA 300
> dre:550447 osgep, si:ch211-214j24.11, wu:fj38e02, zgc:112527;
O-sialoglycoprotein endopeptidase (EC:3.4.24.57); K01409 O-sialoglycoprotein
endopeptidase [EC:3.4.24.57]
Length=335
Score = 106 bits (265), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 50/88 (56%), Positives = 65/88 (73%), Gaps = 0/88 (0%)
Query 40 ESADSSIPEELLTPESICYSLQEYLFGMLVEITERAMALSHSSDVLVVGGVGCNLRLQQM 99
++A + + TPE +C+SLQE +F MLVEITERAMA S +VL+VGGVGCNLRLQ+M
Sbjct 213 DAAHKMLSTDQCTPEDLCFSLQETVFAMLVEITERAMAHCGSQEVLIVGGVGCNLRLQEM 272
Query 100 LREMARQRGCSMGGMDERYCIDNGAMIA 127
+ M ++RG + DE +CIDNGAMIA
Sbjct 273 MGVMCKERGARIFATDESFCIDNGAMIA 300
> hsa:55644 OSGEP, FLJ20411, GCPL1, KAE1, OSGEP1, PRSMG1; O-sialoglycoprotein
endopeptidase (EC:3.4.24.57); K01409 O-sialoglycoprotein
endopeptidase [EC:3.4.24.57]
Length=335
Score = 106 bits (264), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 48/76 (63%), Positives = 60/76 (78%), Gaps = 0/76 (0%)
Query 52 TPESICYSLQEYLFGMLVEITERAMALSHSSDVLVVGGVGCNLRLQQMLREMARQRGCSM 111
TPE +C+SLQE +F MLVEITERAMA S + L+VGGVGCN+RLQ+M+ M ++RG +
Sbjct 225 TPEDLCFSLQETVFAMLVEITERAMAHCGSQEALIVGGVGCNVRLQEMMATMCQERGARL 284
Query 112 GGMDERYCIDNGAMIA 127
DER+CIDNGAMIA
Sbjct 285 FATDERFCIDNGAMIA 300
> sce:YKR038C KAE1; Highly conserved putative glycoprotease proposed
to be involved in transcription as a component of the
EKC protein complex with Bud32p, Cgi121p, Pcc1p, and Gon7p;
also identified as a component of the KEOPS protein complex
(EC:3.4.24.-); K01409 O-sialoglycoprotein endopeptidase [EC:3.4.24.57]
Length=386
Score = 102 bits (254), Expect = 3e-22, Method: Composition-based stats.
Identities = 49/86 (56%), Positives = 63/86 (73%), Gaps = 1/86 (1%)
Query 43 DSSIPEELLTPESICYSLQEYLFGMLVEITERAMALSHSSDVLVVGGVGCNLRLQQMLRE 102
D + E+ +T E +CYSLQE LF MLVEITERAMA +S+ VL+VGGVGCN+RLQ+M+ +
Sbjct 265 DKTTGEQKVTVEDLCYSLQENLFAMLVEITERAMAHVNSNQVLIVGGVGCNVRLQEMMAQ 324
Query 103 MARQRG-CSMGGMDERYCIDNGAMIA 127
M + R + D R+CIDNG MIA
Sbjct 325 MCKDRANGQVHATDNRFCIDNGVMIA 350
> cpv:cgd6_3550 endopeptidase
Length=350
Score = 102 bits (253), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 44/75 (58%), Positives = 60/75 (80%), Gaps = 0/75 (0%)
Query 54 ESICYSLQEYLFGMLVEITERAMALSHSSDVLVVGGVGCNLRLQQMLREMARQRGCSMGG 113
+ C+SLQE LF ML+E+TERA++L +S +L+VGGVGCNLRL +M+ +MA+ RG +
Sbjct 242 QDFCFSLQETLFAMLIEVTERAISLLNSDSILLVGGVGCNLRLIEMMEQMAKDRGAIVCS 301
Query 114 MDERYCIDNGAMIAY 128
MD+ YCIDNGAMIA+
Sbjct 302 MDDSYCIDNGAMIAH 316
> bbo:BBOV_II003000 18.m06249; glycoprotease family protein; K01409
O-sialoglycoprotein endopeptidase [EC:3.4.24.57]
Length=358
Score = 102 bits (253), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 53/113 (46%), Positives = 75/113 (66%), Gaps = 13/113 (11%)
Query 16 EEMAKRMRTKRSGGQLRKRGNPSRESADSSIPEELLTPESICYSLQEYLFGMLVEITERA 75
E++ +R R S G++ + + SR + D +C+S+QE++F ML+E+TERA
Sbjct 222 EQLIERERNLLSSGEITES-DFSRFTCD------------LCFSVQEHMFAMLIEMTERA 268
Query 76 MALSHSSDVLVVGGVGCNLRLQQMLREMARQRGCSMGGMDERYCIDNGAMIAY 128
M+ ++++LVVGGVGCNLRLQ M MA RG + MDERYCIDNGAMIA+
Sbjct 269 MSFVGANELLVVGGVGCNLRLQSMASAMAESRGARLYPMDERYCIDNGAMIAF 321
> tpv:TP04_0293 hypothetical protein; K01409 O-sialoglycoprotein
endopeptidase [EC:3.4.24.57]
Length=363
Score = 100 bits (248), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 47/88 (53%), Positives = 64/88 (72%), Gaps = 1/88 (1%)
Query 41 SADSSIPEELLTPESICYSLQEYLFGMLVEITERAMALSHSSDVLVVGGVGCNLRLQQML 100
S DS+ E + +C+S+QE+ F ML+E+ ERAMA + S ++L+VGGVGCNLRLQ+M
Sbjct 239 SEDSAFEYEQFKVD-LCFSIQEHTFAMLLEMLERAMAFTGSDEILLVGGVGCNLRLQEMA 297
Query 101 REMARQRGCSMGGMDERYCIDNGAMIAY 128
MA++R + MD+RYCIDNGAMI Y
Sbjct 298 NLMAQERNAKLFPMDDRYCIDNGAMIGY 325
> eco:b3064 gcp, ECK3054, JW3036, ygjD; glycation-binding protein,
predicted protease/chaperone; essential for genome maintenance;
K01409 O-sialoglycoprotein endopeptidase [EC:3.4.24.57]
Length=337
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 38/73 (52%), Gaps = 0/73 (0%)
Query 56 ICYSLQEYLFGMLVEITERAMALSHSSDVLVVGGVGCNLRLQQMLREMARQRGCSMGGMD 115
I + ++ + L+ +RA+ + +++ GGV N L+ L EM ++R +
Sbjct 235 IARAFEDAVVDTLMIKCKRALDQTGFKRLVMAGGVSANRTLRAKLAEMMKKRRGEVFYAR 294
Query 116 ERYCIDNGAMIAY 128
+C DNGAMIAY
Sbjct 295 PEFCTDNGAMIAY 307
> mmu:72085 Osgepl1, 2610001M19Rik, AA416452, MGC13061; O-sialoglycoprotein
endopeptidase-like 1 (EC:3.4.24.57); K01409 O-sialoglycoprotein
endopeptidase [EC:3.4.24.57]
Length=414
Score = 37.4 bits (85), Expect = 0.013, Method: Composition-based stats.
Identities = 22/52 (42%), Positives = 31/52 (59%), Gaps = 1/52 (1%)
Query 78 LSHSSDVLVV-GGVGCNLRLQQMLREMARQRGCSMGGMDERYCIDNGAMIAY 128
LS ++ VLVV GGV NL +++ L +A C++ R C DNG MIA+
Sbjct 314 LSPANAVLVVSGGVASNLYIRKALEIVANATQCTLLCPPPRLCTDNGIMIAW 365
> pfa:PFD0440w peptidase, M22 family, putative
Length=693
Score = 36.2 bits (82), Expect = 0.024, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 31/54 (57%), Gaps = 4/54 (7%)
Query 58 YSLQEYLFGMLVEITERAMALS----HSSDVLVVGGVGCNLRLQQMLREMARQR 107
Y Q+ +F L++ + M S + +V +VGGVGCN L Q L++MA +R
Sbjct 465 YYCQKNIFHHLLKQVNKIMYFSELHFNIKNVFIVGGVGCNNFLYQSLKDMAAKR 518
> hsa:64172 OSGEPL1; O-sialoglycoprotein endopeptidase-like 1
(EC:3.4.24.57); K01409 O-sialoglycoprotein endopeptidase [EC:3.4.24.57]
Length=414
Score = 36.2 bits (82), Expect = 0.026, Method: Composition-based stats.
Identities = 24/87 (27%), Positives = 38/87 (43%), Gaps = 8/87 (9%)
Query 50 LLTPESICYSLQEYLFGMLVEITERAMALSHSSDVL--------VVGGVGCNLRLQQMLR 101
L + I ++Q + LV+ T RA+ D+L GGV N +++ L
Sbjct 279 LSSAADIAATVQHTMACHLVKRTHRAILFCKQRDLLPQNNAVLVASGGVASNFYIRRALE 338
Query 102 EMARQRGCSMGGMDERYCIDNGAMIAY 128
+ C++ R C DNG MIA+
Sbjct 339 ILTNATQCTLLCPPPRLCTDNGIMIAW 365
> dre:368635 osgepl1, MGC123084, si:dz72b14.6; O-sialoglycoprotein
endopeptidase-like 1 (EC:3.4.24.57); K01409 O-sialoglycoprotein
endopeptidase [EC:3.4.24.57]
Length=404
Score = 33.1 bits (74), Expect = 0.21, Method: Composition-based stats.
Identities = 23/83 (27%), Positives = 38/83 (45%), Gaps = 8/83 (9%)
Query 54 ESICYSLQEYLFGMLVEITERAMALSHSSDVL--------VVGGVGCNLRLQQMLREMAR 105
+ I + Q + L + T RA+ S +L V GGV N ++Q+L+ +
Sbjct 271 KDIAAASQHTVASHLAKRTHRAILFCKSKGLLPEQNPTLIVSGGVASNEYIRQILKIITD 330
Query 106 QRGCSMGGMDERYCIDNGAMIAY 128
G + ++C DNG MIA+
Sbjct 331 ATGLHLLCPPSKFCTDNGVMIAW 353
> dre:337167 tsc1a, fa99f04, wu:fa99f04, zgc:63657; tuberous sclerosis
1a; K07206 tuberous sclerosis 1
Length=1128
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 28/61 (45%), Gaps = 8/61 (13%)
Query 24 TKRSGGQLRKRGNPSRESADSSIPEELLTPESICYSLQEYLFGMLVEITERAMALSHSSD 83
T+ SG + N S E S + EEL +P SIC GM + R M+ + SD
Sbjct 325 TQDSGSSAQTPNNQSDEVKSSGMKEELWSPSSIC--------GMSTPPSSRGMSPTTVSD 376
Query 84 V 84
+
Sbjct 377 I 377
> tgo:TGME49_074110 hypothetical protein
Length=1323
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 27/58 (46%), Gaps = 5/58 (8%)
Query 56 ICYSLQEYLFGMLVEITERAMAL-----SHSSDVLVVGGVGCNLRLQQMLREMARQRG 108
SLQ +F L + + M L + VVGGV CN L++ LR++ RG
Sbjct 803 FAASLQAAVFKHLEDQLRKTMWLYEFLEDFPRRLAVVGGVSCNETLRRRLRKLCESRG 860
> dre:100007819 solute carrier family 22 member 6-like
Length=520
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 24/97 (24%), Positives = 45/97 (46%), Gaps = 8/97 (8%)
Query 12 PEVAEEMAKRMRTKRSGGQLRKRGNPSRESADSSIPEELLTPESICYSLQEYLFG----- 66
PE A + R RTK + ++K ++ + S+ EE+L + + + LFG
Sbjct 271 PESARWLLDRGRTKDAKKLIQKAAAVNKRAVPESLVEEVLKEKPVEKGGIKILFGSRVLR 330
Query 67 ---MLVEITERAMALSHSSDVLVVGGVGCNLRLQQML 100
+ + A+ L++ S L VG G ++ L Q++
Sbjct 331 KYFLAITFAWCALNLAYYSLSLNVGKFGLDIFLTQLI 367
> mmu:11733 Ank1, Ank-1, nb, pale; ankyrin 1, erythroid; K10380
ankyrin
Length=1907
Score = 28.1 bits (61), Expect = 6.9, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 28/48 (58%), Gaps = 0/48 (0%)
Query 13 EVAEEMAKRMRTKRSGGQLRKRGNPSRESADSSIPEELLTPESICYSL 60
E + ++ ++ +R G +PS+ + SS+ +ELL+P S+ Y+L
Sbjct 1525 EGSGRQSRNLKPERRHGDREYSLSPSQVNGYSSLQDELLSPASLQYAL 1572
> cel:Y105E8A.26 zoo-1; ZO-1 (Zonula Occludens tight junctional
protein) Ortholog family member (zoo-1)
Length=1172
Score = 28.1 bits (61), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 32/71 (45%), Gaps = 0/71 (0%)
Query 13 EVAEEMAKRMRTKRSGGQLRKRGNPSRESADSSIPEELLTPESICYSLQEYLFGMLVEIT 72
E A + + +R + G R + SRE D + P EL E + + + LFG V +
Sbjct 410 EHARQDFEHVRANQLGDNFYIRSHFSREKRDKASPLELSINEGDIFHVTDTLFGGTVGLW 469
Query 73 ERAMALSHSSD 83
+ A S S +
Sbjct 470 QAARVYSSSEN 480
> tgo:TGME49_009860 nucleolar phosphoprotein p130, putative
Length=406
Score = 28.1 bits (61), Expect = 7.5, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 24/46 (52%), Gaps = 2/46 (4%)
Query 4 ALRLTADGPEVAEEMAKRMRTKRSGGQLRKRGNPSRESADSSIPEE 49
AL +AD PE A E K+ + R G+ ++R RE S+ EE
Sbjct 246 ALAGSADAPEAAGEREKKQKKNRFEGKTKQRAQ--REETPSADSEE 289
> mmu:109168 Atl3, 4633402C03Rik, 5730596K20Rik, AI465397, AW228836,
MGC28761; atlastin GTPase 3
Length=536
Score = 27.7 bits (60), Expect = 9.1, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 34/74 (45%), Gaps = 10/74 (13%)
Query 54 ESICYSLQEYLFGMLVE---ITERAMALSHSSDVLVVGGVGCNLRLQQMLREMARQRGCS 110
E IC + YL ++E + + +AL H + +GG + R QQ L E ++
Sbjct 363 EEICGGEKPYLSPDILEEKHLEFKQLALDHFKKIKKMGGKDFSFRYQQELEEEIKE---- 418
Query 111 MGGMDERYCIDNGA 124
+ E +C NG+
Sbjct 419 ---LYENFCKHNGS 429
Lambda K H
0.319 0.133 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2049573556
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40