bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5083_orf2
Length=91
Score E
Sequences producing significant alignments: (Bits) Value
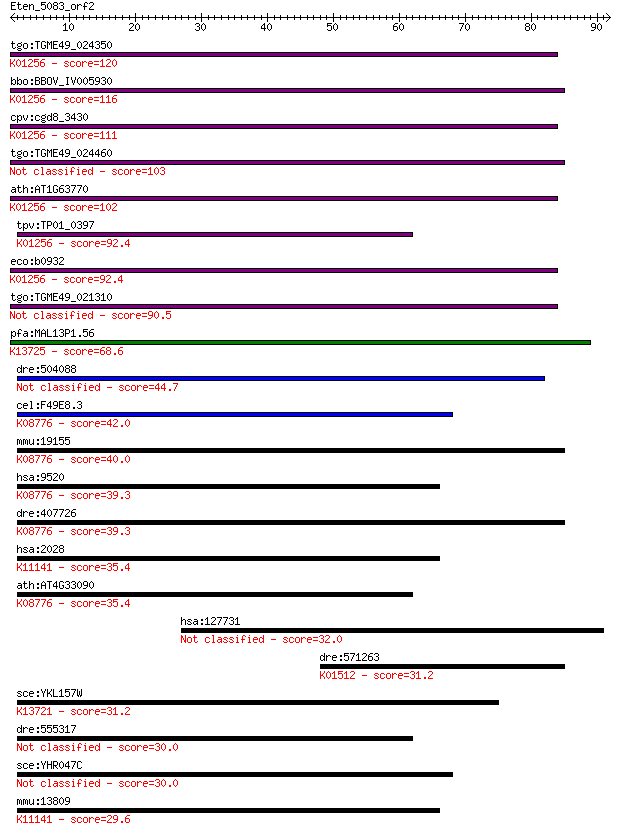
tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2); K0... 120 9e-28
bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01... 116 1e-26
cpv:cgd8_3430 zincin/aminopeptidase N like metalloprotease ; K... 111 6e-25
tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2) 103 1e-22
ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptida... 102 3e-22
tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2); ... 92.4 3e-19
eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2... 92.4 3e-19
tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2) 90.5 1e-18
pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725 ... 68.6 5e-12
dre:504088 enpep, im:7152184, si:ch211-146m5.2; glutamyl amino... 44.7 7e-05
cel:F49E8.3 pam-1; Puromycin-sensitive AMinopeptidase family m... 42.0 5e-04
mmu:19155 Npepps, AAP-S, MGC102199, MP100, Psa, R74825, goku; ... 40.0 0.002
hsa:9520 NPEPPS, AAP-S, MP100, PSA; aminopeptidase puromycin s... 39.3 0.003
dre:407726 npepps, Psa, fb68d07, sb:cb848, wu:fb68d07; aminope... 39.3 0.003
hsa:2028 ENPEP, APA, CD249, gp160; glutamyl aminopeptidase (am... 35.4 0.044
ath:AT4G33090 APM1; APM1 (AMINOPEPTIDASE M1); aminopeptidase; ... 35.4 0.045
hsa:127731 VWA5B1, FLJ32784, FLJ43845; von Willebrand factor A... 32.0 0.46
dre:571263 acyp1, MGC163122, eap1, im:7149755, wu:fj62c08, zgc... 31.2 0.84
sce:YKL157W APE2, LAP1, YKL158W; Aminopeptidase yscII; may hav... 31.2 0.86
dre:555317 aminopeptidase N-like 30.0 1.6
sce:YHR047C AAP1, AAP1'; Aap1p (EC:3.4.11.-) 30.0 1.9
mmu:13809 Enpep, 6030431M22Rik, APA, Bp-1/6C3, Ly-51, Ly51; gl... 29.6 2.6
> tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1419
Score = 120 bits (302), Expect = 9e-28, Method: Composition-based stats.
Identities = 53/83 (63%), Positives = 64/83 (77%), Gaps = 0/83 (0%)
Query 1 EVVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNLDQMERWYTQAGT 60
EV+R+YHTLLG GFRKGMDLYF+RHD +A TCDDFRAAMAD+NG +L Q ERWY QAGT
Sbjct 924 EVIRMYHTLLGEAGFRKGMDLYFKRHDGKAVTCDDFRAAMADANGRDLGQFERWYLQAGT 983
Query 61 PRLEVXKAALNKETKVFEIRFKQ 83
P + V +A + K F++ KQ
Sbjct 984 PEVTVSEAVFQPDRKKFKLTLKQ 1006
> bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01256
aminopeptidase N [EC:3.4.11.2]
Length=846
Score = 116 bits (291), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 53/84 (63%), Positives = 65/84 (77%), Gaps = 2/84 (2%)
Query 1 EVVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNLDQMERWYTQAGT 60
EV+ +Y TLLG GFRKGMDLYF+RHD+ A TCDDFRAAMAD+NGV+L Q ERWY QAGT
Sbjct 423 EVIGMYKTLLGKDGFRKGMDLYFERHDSHAVTCDDFRAAMADANGVDLTQFERWYFQAGT 482
Query 61 PRLEVXKAALNKETKVFEIRFKQY 84
P +EV +A ++ F +R +QY
Sbjct 483 PEVEVLEAV--RDGTTFRLRLRQY 504
> cpv:cgd8_3430 zincin/aminopeptidase N like metalloprotease ;
K01256 aminopeptidase N [EC:3.4.11.2]
Length=936
Score = 111 bits (277), Expect = 6e-25, Method: Composition-based stats.
Identities = 50/83 (60%), Positives = 55/83 (66%), Gaps = 0/83 (0%)
Query 1 EVVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNLDQMERWYTQAGT 60
EVVR+Y T+LG GFRKGMDLYF RHD QA TCDDFR AM D+N N Q ERWY QAGT
Sbjct 425 EVVRMYETILGREGFRKGMDLYFARHDGQAVTCDDFRKAMEDANNYNFTQFERWYDQAGT 484
Query 61 PRLEVXKAALNKETKVFEIRFKQ 83
P +EV NK I +Q
Sbjct 485 PEVEVVSIDHNKAEGTCSITLRQ 507
> tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2)
Length=970
Score = 103 bits (257), Expect = 1e-22, Method: Composition-based stats.
Identities = 45/84 (53%), Positives = 58/84 (69%), Gaps = 0/84 (0%)
Query 1 EVVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNLDQMERWYTQAGT 60
EV R+Y T+LG GFR+GMDLYF+RHD +A TCDD RAAMAD+N +L Q ERWY+QAGT
Sbjct 465 EVNRMYRTMLGPKGFRRGMDLYFKRHDGKAVTCDDLRAAMADANDKDLSQFERWYSQAGT 524
Query 61 PRLEVXKAALNKETKVFEIRFKQY 84
P + V + + + KQ+
Sbjct 525 PHVTVSSFIYDAAERKMHLTLKQH 548
> ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptidase
N [EC:3.4.11.2]
Length=987
Score = 102 bits (254), Expect = 3e-22, Method: Composition-based stats.
Identities = 45/83 (54%), Positives = 61/83 (73%), Gaps = 1/83 (1%)
Query 1 EVVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNLDQMERWYTQAGT 60
EVVR+Y TLLGT GFRKG+DLYF+RHD QA TC+DF AAM D+N + +WY+QAGT
Sbjct 494 EVVRMYKTLLGTQGFRKGIDLYFERHDEQAVTCEDFFAAMRDANNADFANFLQWYSQAGT 553
Query 61 PRLEVXKAALNKETKVFEIRFKQ 83
P ++V ++ N + + F ++F Q
Sbjct 554 PVVKVV-SSYNADARTFSLKFSQ 575
> tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1020
Score = 92.4 bits (228), Expect = 3e-19, Method: Composition-based stats.
Identities = 39/60 (65%), Positives = 47/60 (78%), Gaps = 0/60 (0%)
Query 2 VVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNLDQMERWYTQAGTP 61
V+ +Y +LLG GFR+GMDLYF+RHD A TCDDFR AMAD+N +L Q ERWY Q+GTP
Sbjct 546 VIGMYESLLGVDGFRRGMDLYFKRHDLSAVTCDDFRLAMADANNKDLTQFERWYYQSGTP 605
> eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=870
Score = 92.4 bits (228), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 43/83 (51%), Positives = 56/83 (67%), Gaps = 1/83 (1%)
Query 1 EVVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNLDQMERWYTQAGT 60
EV+R+ HTLLG F+KGM LYF+RHD AATCDDF AM D++ V+L RWY+Q+GT
Sbjct 386 EVIRMIHTLLGEENFQKGMQLYFERHDGSAATCDDFVQAMEDASNVDLSHFRRWYSQSGT 445
Query 61 PRLEVXKAALNKETKVFEIRFKQ 83
P + V K N ET+ + + Q
Sbjct 446 PIVTV-KDDYNPETEQYTLTISQ 467
> tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2)
Length=966
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 40/83 (48%), Positives = 55/83 (66%), Gaps = 0/83 (0%)
Query 1 EVVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNLDQMERWYTQAGT 60
E+ R+Y TLLG FRKG++LYF R+D QAATC++FR+AM +++G NL Q WYT+ GT
Sbjct 435 EIFRMYATLLGPSAFRKGLNLYFSRYDGQAATCENFRSAMEEASGRNLSQFFLWYTREGT 494
Query 61 PRLEVXKAALNKETKVFEIRFKQ 83
P +E+ +K K F Q
Sbjct 495 PEVEITGFTFDKTRKQFSFTVTQ 517
> pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725
M1-family aminopeptidase [EC:3.4.11.-]
Length=1085
Score = 68.6 bits (166), Expect = 5e-12, Method: Composition-based stats.
Identities = 35/95 (36%), Positives = 53/95 (55%), Gaps = 8/95 (8%)
Query 1 EVVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADS-------NGVNLDQMER 53
EV+R+Y T+LG ++KG D+Y +++D ATC+DF AM + N NL+Q
Sbjct 585 EVMRMYLTILGEEYYKKGFDIYIKKNDGNTATCEDFNYAMEQAYKMKKADNSANLNQYLL 644
Query 54 WYTQAGTPRLEVXKAALNKETKVFEIRFKQYLGRD 88
W++Q+GTP + K + E K + I QY D
Sbjct 645 WFSQSGTPHVSF-KYNYDAEKKQYSIHVNQYTKPD 678
> dre:504088 enpep, im:7152184, si:ch211-146m5.2; glutamyl aminopeptidase
(EC:3.4.11.7)
Length=951
Score = 44.7 bits (104), Expect = 7e-05, Method: Composition-based stats.
Identities = 27/81 (33%), Positives = 41/81 (50%), Gaps = 3/81 (3%)
Query 2 VVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNL-DQMERWYTQAGT 60
++R+ LLG FR G Y + + Q A DF A+AD +G+ + D M+ W Q G
Sbjct 476 ILRMLEDLLGRETFRDGCRRYLKTYLFQNAKTSDFWKALADESGLPVADIMDTWTKQMGY 535
Query 61 PRLEVXKAALNKETKVFEIRF 81
P L + + E K+ + RF
Sbjct 536 PVLSLTNT--DTEAKLTQTRF 554
> cel:F49E8.3 pam-1; Puromycin-sensitive AMinopeptidase family
member (pam-1); K08776 puromycin-sensitive aminopeptidase [EC:3.4.11.-]
Length=948
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 36/67 (53%), Gaps = 1/67 (1%)
Query 2 VVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNLDQ-MERWYTQAGT 60
V R+ L P F+KG+ LY +R A D A+++++G N+++ M W Q G
Sbjct 469 VNRMLCYYLSEPVFQKGLRLYLKRFQYSNAVTQDLWTALSEASGQNVNELMSGWTQQMGF 528
Query 61 PRLEVXK 67
P L+V +
Sbjct 529 PVLKVSQ 535
> mmu:19155 Npepps, AAP-S, MGC102199, MP100, Psa, R74825, goku;
aminopeptidase puromycin sensitive (EC:3.4.11.14); K08776
puromycin-sensitive aminopeptidase [EC:3.4.11.-]
Length=920
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 21/84 (25%), Positives = 44/84 (52%), Gaps = 2/84 (2%)
Query 2 VVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNLDQ-MERWYTQAGT 60
V+R+ H +G F+KGM++Y + + A +D ++ ++G + M W Q G
Sbjct 445 VIRMLHDYIGDKDFKKGMNMYLTKFQQKNAATEDLWESLESASGKPIAAVMNTWTKQMGF 504
Query 61 PRLEVXKAALNKETKVFEIRFKQY 84
P + V +A ++ +V ++ K++
Sbjct 505 PLIYV-EAEQVEDDRVLKLSQKKF 527
> hsa:9520 NPEPPS, AAP-S, MP100, PSA; aminopeptidase puromycin
sensitive (EC:3.4.11.14); K08776 puromycin-sensitive aminopeptidase
[EC:3.4.11.-]
Length=919
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 34/65 (52%), Gaps = 1/65 (1%)
Query 2 VVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNLDQ-MERWYTQAGT 60
V+R+ H +G F+KGM++Y + + A +D ++ +++G + M W Q G
Sbjct 444 VIRMLHDYIGDKDFKKGMNMYLTKFQQKNAATEDLWESLENASGKPIAAVMNTWTKQMGF 503
Query 61 PRLEV 65
P + V
Sbjct 504 PLIYV 508
> dre:407726 npepps, Psa, fb68d07, sb:cb848, wu:fb68d07; aminopeptidase
puromycin sensitive (EC:3.4.11.-); K08776 puromycin-sensitive
aminopeptidase [EC:3.4.11.-]
Length=872
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 22/84 (26%), Positives = 42/84 (50%), Gaps = 2/84 (2%)
Query 2 VVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNLDQ-MERWYTQAGT 60
V+R+ H +G FRKGM+ Y + ++ A+ +D + ++G + M W Q G
Sbjct 399 VIRMLHNYIGDEDFRKGMNAYLLKFQHKNASTEDLWECLEQASGKPIAAVMNSWTKQMGF 458
Query 61 PRLEVXKAALNKETKVFEIRFKQY 84
P + V + + +V +I K++
Sbjct 459 PIIVVDQEQHGSD-RVLKISQKKF 481
> hsa:2028 ENPEP, APA, CD249, gp160; glutamyl aminopeptidase (aminopeptidase
A) (EC:3.4.11.7); K11141 glutamyl aminopeptidase
[EC:3.4.11.7]
Length=957
Score = 35.4 bits (80), Expect = 0.044, Method: Composition-based stats.
Identities = 17/65 (26%), Positives = 35/65 (53%), Gaps = 1/65 (1%)
Query 2 VVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNLDQ-MERWYTQAGT 60
++R+ + F+KG +Y +++ + A DF AA+ +++ + + + M+ W Q G
Sbjct 485 ILRMLEDWIKPENFQKGCQMYLEKYQFKNAKTSDFWAALEEASRLPVKEVMDTWTRQMGY 544
Query 61 PRLEV 65
P L V
Sbjct 545 PVLNV 549
> ath:AT4G33090 APM1; APM1 (AMINOPEPTIDASE M1); aminopeptidase;
K08776 puromycin-sensitive aminopeptidase [EC:3.4.11.-]
Length=879
Score = 35.4 bits (80), Expect = 0.045, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 30/61 (49%), Gaps = 1/61 (1%)
Query 2 VVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNLDQ-MERWYTQAGT 60
V+R+ + LG F+K + Y + H A +D AA+ +G +++ M W Q G
Sbjct 398 VIRMLQSYLGAEVFQKSLAAYIKNHAYSNAKTEDLWAALEAGSGEPVNKLMSSWTKQKGY 457
Query 61 P 61
P
Sbjct 458 P 458
> hsa:127731 VWA5B1, FLJ32784, FLJ43845; von Willebrand factor
A domain containing 5B1
Length=1215
Score = 32.0 bits (71), Expect = 0.46, Method: Composition-based stats.
Identities = 23/70 (32%), Positives = 31/70 (44%), Gaps = 12/70 (17%)
Query 27 DNQAATCDDFRAAMADSNGVNLDQMERWYTQA----GTPRL--EVXKAALNKETKVFEIR 80
D+ A CDD + AD G N+ +W + G PRL + A+N KV E
Sbjct 419 DSLAMACDDIQRMKADMGGTNILSPLKWVIRQPVHRGHPRLLFVITDGAVNNTGKVLE-- 476
Query 81 FKQYLGRDHA 90
L R+HA
Sbjct 477 ----LVRNHA 482
> dre:571263 acyp1, MGC163122, eap1, im:7149755, wu:fj62c08, zgc:163122;
acylphosphatase 1, erythrocyte (common) type (EC:3.6.1.7);
K01512 acylphosphatase [EC:3.6.1.7]
Length=99
Score = 31.2 bits (69), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 11/37 (29%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 48 LDQMERWYTQAGTPRLEVXKAALNKETKVFEIRFKQY 84
+ QM++W G+P+ + KA E + E+ FK +
Sbjct 59 VQQMQQWLQTTGSPKSRIAKAEFQNEHPIHELEFKDF 95
> sce:YKL157W APE2, LAP1, YKL158W; Aminopeptidase yscII; may have
a role in obtaining leucine from dipeptide substrates; sequence
coordinates have changed since RT-PCR analysis showed
that the adjacent ORF YKL158W comprises the 5' exon of APE2/YKL157W;
K13721 aminopeptidase 2 [EC:3.4.11.-]
Length=935
Score = 31.2 bits (69), Expect = 0.86, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 34/74 (45%), Gaps = 1/74 (1%)
Query 2 VVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNLDQ-MERWYTQAGT 60
++R+ LG F KG+ Y + A +D A+AD++G ++ M W + G
Sbjct 488 LLRMISKWLGEETFIKGVSQYLNKFKYGNAKTEDLWDALADASGKDVRSVMNIWTKKVGF 547
Query 61 PRLEVXKAALNKET 74
P + V + K T
Sbjct 548 PVISVSEDGNGKIT 561
> dre:555317 aminopeptidase N-like
Length=956
Score = 30.0 bits (66), Expect = 1.6, Method: Composition-based stats.
Identities = 23/69 (33%), Positives = 30/69 (43%), Gaps = 9/69 (13%)
Query 2 VVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAM---ADSNG-VNL-----DQME 52
V+R+ L F KG+ Y Q H D + ADS+G V+L + M
Sbjct 479 VLRMLSEFLSESVFAKGLHNYLQEHAYSNTVYTDLWKKLQEVADSDGNVHLPASIEEIMN 538
Query 53 RWYTQAGTP 61
RW QAG P
Sbjct 539 RWILQAGFP 547
> sce:YHR047C AAP1, AAP1'; Aap1p (EC:3.4.11.-)
Length=856
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 31/67 (46%), Gaps = 1/67 (1%)
Query 2 VVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNL-DQMERWYTQAGT 60
++R+ LG F KG+ Y + A D A+AD++G ++ M W + G
Sbjct 392 LLRMISKWLGEETFIKGVSQYLNKFKYGNAKTGDLWDALADASGKDVCSVMNIWTKRVGF 451
Query 61 PRLEVXK 67
P L V +
Sbjct 452 PVLSVKE 458
> mmu:13809 Enpep, 6030431M22Rik, APA, Bp-1/6C3, Ly-51, Ly51;
glutamyl aminopeptidase (EC:3.4.11.7); K11141 glutamyl aminopeptidase
[EC:3.4.11.7]
Length=945
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 14/65 (21%), Positives = 33/65 (50%), Gaps = 1/65 (1%)
Query 2 VVRIYHTLLGTPGFRKGMDLYFQRHDNQAATCDDFRAAMADSNGVNLDQ-MERWYTQAGT 60
++R+ + F+KG +Y ++ A DF ++ +++ + + + M+ W +Q G
Sbjct 477 ILRMLQDWITPEKFQKGCQIYLKKFQFANAKTSDFWDSLQEASNLPVKEVMDTWTSQMGY 536
Query 61 PRLEV 65
P + V
Sbjct 537 PVVTV 541
Lambda K H
0.322 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2007827920
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40