bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5089_orf2
Length=80
Score E
Sequences producing significant alignments: (Bits) Value
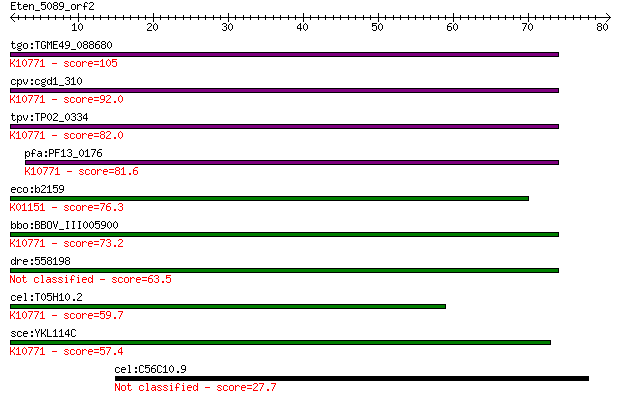
tgo:TGME49_088680 endonuclease V, putative (EC:3.1.21.2); K107... 105 3e-23
cpv:cgd1_310 AP endonuclease of the TIM barrel fold, possible ... 92.0 4e-19
tpv:TP02_0334 apurinic/apyrimidinic endonuclease; K10771 AP en... 82.0 4e-16
pfa:PF13_0176 apn1; apurinic/apyrimidinic endonuclease Apn1 (E... 81.6 5e-16
eco:b2159 nfo, ECK2152, JW2146; endonuclease IV with intrinsic... 76.3 2e-14
bbo:BBOV_III005900 17.m07523; apurinic endonuclease (APN1) fam... 73.2 2e-13
dre:558198 APurinic/apyrimidinic endoNuclease family member (a... 63.5 2e-10
cel:T05H10.2 apn-1; APurinic/apyrimidinic endoNuclease family ... 59.7 2e-09
sce:YKL114C APN1; Apn1p (EC:4.2.99.18); K10771 AP endonuclease... 57.4 1e-08
cel:C56C10.9 hypothetical protein 27.7 8.9
> tgo:TGME49_088680 endonuclease V, putative (EC:3.1.21.2); K10771
AP endonuclease 1 [EC:4.2.99.18]
Length=667
Score = 105 bits (263), Expect = 3e-23, Method: Composition-based stats.
Identities = 48/75 (64%), Positives = 58/75 (77%), Gaps = 2/75 (2%)
Query 1 GYKFLRGMHLNDSKSELSSGLDRHELLGKGHLGLAPFKYIMQHQTRFKDMPLILETPDPT 60
G K+L+ MH+NDSK+ L SGLDRHE LGKG LG+APFK+IMQH T FKDMPL+LETPD
Sbjct 588 GMKYLKAMHINDSKAPLGSGLDRHEHLGKGTLGMAPFKFIMQHPTWFKDMPLVLETPDVD 647
Query 61 EG--TIWKKEIKQMY 73
+W+KE + MY
Sbjct 648 NSGPAMWRKETEMMY 662
> cpv:cgd1_310 AP endonuclease of the TIM barrel fold, possible
bacterial horizontal transfer ; K10771 AP endonuclease 1 [EC:4.2.99.18]
Length=364
Score = 92.0 bits (227), Expect = 4e-19, Method: Composition-based stats.
Identities = 43/74 (58%), Positives = 58/74 (78%), Gaps = 2/74 (2%)
Query 1 GYKFLRGMHLNDSKSELSSGLDRHELLGKGHLGLAPFKYIMQHQTRFKDMPLILETPDP- 59
G K+L+ MHLNDSK + +SGLDRHE LGKG++G+ FK+IM + +RF D+PLILETPDP
Sbjct 285 GLKYLKAMHLNDSKGQFNSGLDRHENLGKGNIGMECFKFIM-NDSRFNDIPLILETPDPN 343
Query 60 TEGTIWKKEIKQMY 73
+ ++KKEI +Y
Sbjct 344 NDDKVYKKEIAILY 357
> tpv:TP02_0334 apurinic/apyrimidinic endonuclease; K10771 AP
endonuclease 1 [EC:4.2.99.18]
Length=422
Score = 82.0 bits (201), Expect = 4e-16, Method: Composition-based stats.
Identities = 42/75 (56%), Positives = 56/75 (74%), Gaps = 4/75 (5%)
Query 1 GYKFLRGMHLNDSKSELSSGLDRHELLGKGHLGLAPFKYIMQHQTRFKDMPLILETPDPT 60
G K+LR +HLNDSKS+L SGLDRHE +GKG L F++I+ + FK+MP+ILETP P
Sbjct 346 GLKYLRAVHLNDSKSDLGSGLDRHENIGKGKLSEQTFRFIV-NSPYFKNMPIILETPVP- 403
Query 61 EGT--IWKKEIKQMY 73
EG ++K+E+K MY
Sbjct 404 EGNEEVYKQEVKLMY 418
> pfa:PF13_0176 apn1; apurinic/apyrimidinic endonuclease Apn1
(EC:4.2.99.18); K10771 AP endonuclease 1 [EC:4.2.99.18]
Length=597
Score = 81.6 bits (200), Expect = 5e-16, Method: Composition-based stats.
Identities = 40/72 (55%), Positives = 57/72 (79%), Gaps = 2/72 (2%)
Query 3 KFLRGMHLNDSKSELSSGLDRHELLGKGHLGLAPFKYIMQHQTRFKDMPLILETPDPT-E 61
K+L+ +HLNDSKS++ SGLDRHE +GKG L + FKYIM+ + FK++P+ILETPD T +
Sbjct 521 KYLKAVHLNDSKSDIGSGLDRHENIGKGKLTMDTFKYIMKSKY-FKNIPIILETPDITND 579
Query 62 GTIWKKEIKQMY 73
+I+K EI+ +Y
Sbjct 580 ESIYKYEIQNLY 591
> eco:b2159 nfo, ECK2152, JW2146; endonuclease IV with intrinsic
3'-5' exonuclease activity (EC:3.1.21.2); K01151 deoxyribonuclease
IV [EC:3.1.21.2]
Length=285
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 37/69 (53%), Positives = 49/69 (71%), Gaps = 4/69 (5%)
Query 1 GYKFLRGMHLNDSKSELSSGLDRHELLGKGHLGLAPFKYIMQHQTRFKDMPLILETPDPT 60
G+K+LRGMHLND+KS S +DRH LG+G++G F++IMQ RF +PLILET +P
Sbjct 208 GFKYLRGMHLNDAKSTFGSRVDRHHSLGEGNIGHDAFRWIMQ-DDRFDGIPLILETINP- 265
Query 61 EGTIWKKEI 69
IW +EI
Sbjct 266 --DIWAEEI 272
> bbo:BBOV_III005900 17.m07523; apurinic endonuclease (APN1) family
protein; K10771 AP endonuclease 1 [EC:4.2.99.18]
Length=374
Score = 73.2 bits (178), Expect = 2e-13, Method: Composition-based stats.
Identities = 38/74 (51%), Positives = 52/74 (70%), Gaps = 2/74 (2%)
Query 1 GYKFLRGMHLNDSKSELSSGLDRHELLGKGHLGLAPFKYIMQHQTRFKDMPLILETPD-P 59
G K+L +HLND KS+L SGLDRHE +G G L F++I + F++MP+ILETPD
Sbjct 290 GLKYLVAVHLNDCKSDLGSGLDRHENIGIGKLTRETFEFI-ANSGYFRNMPIILETPDIH 348
Query 60 TEGTIWKKEIKQMY 73
+ TI+K+E+K MY
Sbjct 349 GDETIYKQEVKVMY 362
> dre:558198 APurinic/apyrimidinic endoNuclease family member
(apn-1)-like
Length=296
Score = 63.5 bits (153), Expect = 2e-10, Method: Composition-based stats.
Identities = 32/73 (43%), Positives = 50/73 (68%), Gaps = 2/73 (2%)
Query 1 GYKFLRGMHLNDSKSELSSGLDRHELLGKGHLGLAPFKYIMQHQTRFKDMPLILETPDPT 60
G +LR +HLNDSK +L S LDRHE +G G +G+ F+ I+ ++ R ++PLILETP
Sbjct 217 GLHYLRAVHLNDSKGKLGSHLDRHEDIGHGQIGITAFREIV-NEPRLDNIPLILETPG-R 274
Query 61 EGTIWKKEIKQMY 73
+G + ++I+ +Y
Sbjct 275 QGFEYAEQIELLY 287
> cel:T05H10.2 apn-1; APurinic/apyrimidinic endoNuclease family
member (apn-1); K10771 AP endonuclease 1 [EC:4.2.99.18]
Length=396
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 26/58 (44%), Positives = 42/58 (72%), Gaps = 1/58 (1%)
Query 1 GYKFLRGMHLNDSKSELSSGLDRHELLGKGHLGLAPFKYIMQHQTRFKDMPLILETPD 58
G+ +L+ +H+NDSK ++ S LDRHE +G+G +G A F+ +M + R +P+ILETP+
Sbjct 324 GWNYLKAIHINDSKGDVGSKLDRHEHIGQGKIGKAAFELLM-NDNRLDGIPMILETPE 380
> sce:YKL114C APN1; Apn1p (EC:4.2.99.18); K10771 AP endonuclease
1 [EC:4.2.99.18]
Length=367
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 32/73 (43%), Positives = 47/73 (64%), Gaps = 4/73 (5%)
Query 1 GYKFLRGMHLNDSKSELSSGLDRHELLGKGHLGLAPFKYIMQHQTRFKDMPLILETP-DP 59
G+K+L +HLNDSK+ L + D HE LG+G+LG+ F+ I H + +P++LETP +
Sbjct 221 GFKYLSAVHLNDSKAPLGANRDLHERLGQGYLGIDVFRMIA-HSEYLQGIPIVLETPYEN 279
Query 60 TEGTIWKKEIKQM 72
EG + EIK M
Sbjct 280 DEG--YGNEIKLM 290
> cel:C56C10.9 hypothetical protein
Length=320
Score = 27.7 bits (60), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 16/63 (25%), Positives = 29/63 (46%), Gaps = 0/63 (0%)
Query 15 SELSSGLDRHELLGKGHLGLAPFKYIMQHQTRFKDMPLILETPDPTEGTIWKKEIKQMYL 74
S LS L+R + L H+ L P + + +F+ + DP++ + IK+M+
Sbjct 13 SALSIPLNRSDFLENDHVELMPLEKDGELNDKFRQEMIFSHNLDPSDSKQLSESIKEMFK 72
Query 75 GRD 77
D
Sbjct 73 KTD 75
Lambda K H
0.319 0.139 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2058492688
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40