bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5122_orf1
Length=79
Score E
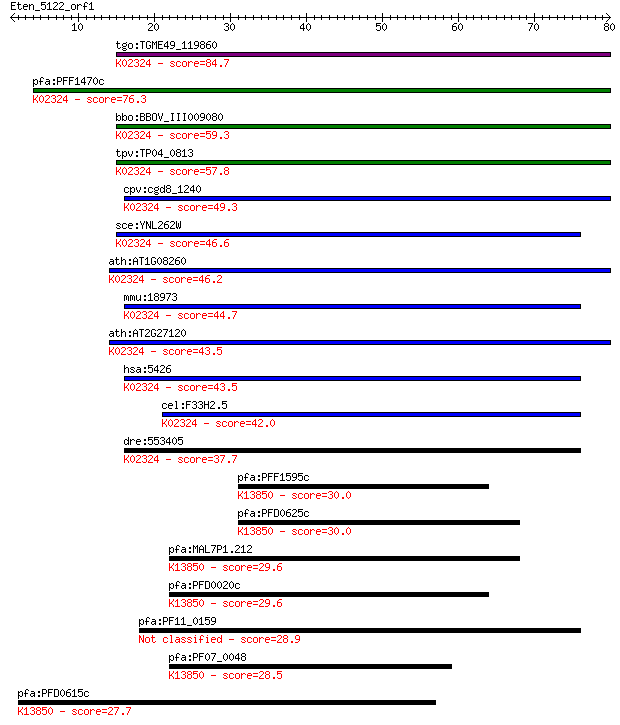
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_119860 DNA polymerase epsilon catalytic subunit, pu... 84.7 6e-17
pfa:PFF1470c DNA polymerase epsilon, catalytic subunit a, puta... 76.3 2e-14
bbo:BBOV_III009080 17.m07793; DNA polymerase family B, exonucl... 59.3 3e-09
tpv:TP04_0813 DNA polymerase epsilon, catalytic subunit A (EC:... 57.8 1e-08
cpv:cgd8_1240 DNA polymerase epsilon catalytic subunit ; K0232... 49.3 3e-06
sce:YNL262W POL2, DUN2; Catalytic subunit of DNA polymerase (I... 46.6 2e-05
ath:AT1G08260 TIL1; TIL1 (TILTED 1); DNA binding / DNA-directe... 46.2 2e-05
mmu:18973 Pole; polymerase (DNA directed), epsilon (EC:2.7.7.7... 44.7 8e-05
ath:AT2G27120 TIL2; TIL2 (TILTED 2); DNA binding / DNA-directe... 43.5 2e-04
hsa:5426 POLE, DKFZp434F222, FLJ21434, POLE1; polymerase (DNA ... 43.5 2e-04
cel:F33H2.5 hypothetical protein; K02324 DNA polymerase epsilo... 42.0 5e-04
dre:553405 pole, si:dkeyp-98c11.2; polymerase (DNA directed), ... 37.7 0.010
pfa:PFF1595c VAR; erythrocyte membrane protein 1, PfEMP1; K138... 30.0 1.7
pfa:PFD0625c VAR; erythrocyte membrane protein 1, PfEMP1; K138... 30.0 1.9
pfa:MAL7P1.212 VAR; erythrocyte membrane protein 1, PfEMP1; K1... 29.6 2.4
pfa:PFD0020c VAR; erythrocyte membrane protein 1, PfEMP1; K138... 29.6 2.5
pfa:PF11_0159 conserved Plasmodium protein 28.9 4.2
pfa:PF07_0048 VAR; erythrocyte membrane protein 1, PfEMP1; K13... 28.5 5.9
pfa:PFD0615c VAR; erythrocyte membrane protein 1, PfEMP1; K138... 27.7 9.0
> tgo:TGME49_119860 DNA polymerase epsilon catalytic subunit,
putative (EC:2.7.11.12 2.7.7.7); K02324 DNA polymerase epsilon
subunit 1 [EC:2.7.7.7]
Length=3124
Score = 84.7 bits (208), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 35/65 (53%), Positives = 50/65 (76%), Gaps = 0/65 (0%)
Query 15 RKLRFEFPTWLLNFAVYRRFRNEMFLSFCPERKSFLKEIKNEILFELDGPWKGMFLPASE 74
++ RFEFPT +LN V+RR+ N+ +L F + ++F ++ +NE+ FELDGPW+ MFLPASE
Sbjct 1245 KEFRFEFPTTILNQDVHRRYTNDQYLDFDEDSRTFKRQTRNEVYFELDGPWRAMFLPASE 1304
Query 75 KSDSL 79
KSD L
Sbjct 1305 KSDDL 1309
> pfa:PFF1470c DNA polymerase epsilon, catalytic subunit a, putative
(EC:2.7.7.7); K02324 DNA polymerase epsilon subunit 1
[EC:2.7.7.7]
Length=2907
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 36/76 (47%), Positives = 49/76 (64%), Gaps = 0/76 (0%)
Query 4 SSSSSSSSRGVRKLRFEFPTWLLNFAVYRRFRNEMFLSFCPERKSFLKEIKNEILFELDG 63
S + V+K+ FEFPT +LNF +++++ N+ +L + + KNEI FELDG
Sbjct 1006 SEEELKNDNNVKKVDFEFPTNILNFEMHKKWTNDQYLVYNEHTDDYECISKNEIFFELDG 1065
Query 64 PWKGMFLPASEKSDSL 79
PW GMFLPASEKSD L
Sbjct 1066 PWHGMFLPASEKSDDL 1081
> bbo:BBOV_III009080 17.m07793; DNA polymerase family B, exonuclease
domain containing protein; K02324 DNA polymerase epsilon
subunit 1 [EC:2.7.7.7]
Length=2346
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 29/65 (44%), Positives = 42/65 (64%), Gaps = 1/65 (1%)
Query 15 RKLRFEFPTWLLNFAVYRRFRNEMFLSFCPERKSFLKEIKNEILFELDGPWKGMFLPASE 74
+ L E+ T +LN + R++ N+ +L + + + +NEILFELDGPW MFLPASE
Sbjct 968 KTLELEYMTTVLNRLIERQWTNDQYLEM-EDIGKYRRVRRNEILFELDGPWHAMFLPASE 1026
Query 75 KSDSL 79
KS+ L
Sbjct 1027 KSEDL 1031
> tpv:TP04_0813 DNA polymerase epsilon, catalytic subunit A (EC:2.7.7.7);
K02324 DNA polymerase epsilon subunit 1 [EC:2.7.7.7]
Length=1430
Score = 57.8 bits (138), Expect = 1e-08, Method: Composition-based stats.
Identities = 28/65 (43%), Positives = 39/65 (60%), Gaps = 1/65 (1%)
Query 15 RKLRFEFPTWLLNFAVYRRFRNEMFLSFCPERKSFLKEIKNEILFELDGPWKGMFLPASE 74
+ + E+ T +LN + + NE +L + ++ KNEI FELDGPW MFLPASE
Sbjct 983 KSVELEYMTTVLNMLIAEEWTNEQYLDL-EDSGRYVTVQKNEIFFELDGPWHAMFLPASE 1041
Query 75 KSDSL 79
KS+ L
Sbjct 1042 KSEEL 1046
> cpv:cgd8_1240 DNA polymerase epsilon catalytic subunit ; K02324
DNA polymerase epsilon subunit 1 [EC:2.7.7.7]
Length=2728
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 39/64 (60%), Gaps = 0/64 (0%)
Query 16 KLRFEFPTWLLNFAVYRRFRNEMFLSFCPERKSFLKEIKNEILFELDGPWKGMFLPASEK 75
K + + LLN V++ + N+ +L + + + +++N I FE+DGP+K F+PAS+K
Sbjct 936 KKKIHYICTLLNQQVHKSWTNDQYLEYDADLGKYNIKVENSIYFEVDGPYKCFFIPASDK 995
Query 76 SDSL 79
+ L
Sbjct 996 QNKL 999
> sce:YNL262W POL2, DUN2; Catalytic subunit of DNA polymerase
(II) epsilon, a chromosomal DNA replication polymerase that
exhibits processivity and proofreading exonuclease activity;
also involved in DNA synthesis during DNA repair; interacts
extensively with Mrc1p (EC:2.7.7.7); K02324 DNA polymerase
epsilon subunit 1 [EC:2.7.7.7]
Length=2222
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 36/62 (58%), Gaps = 1/62 (1%)
Query 15 RKLRFEFPTWLLNFAVYRRFRNEMFLSF-CPERKSFLKEIKNEILFELDGPWKGMFLPAS 73
+KL +P +LN+ V+++F N + P + +N I FE+DGP+K M LP+S
Sbjct 899 KKLYLSYPCSMLNYRVHQKFTNHQYQELKDPLNYIYETHSENTIFFEVDGPYKAMILPSS 958
Query 74 EK 75
++
Sbjct 959 KE 960
> ath:AT1G08260 TIL1; TIL1 (TILTED 1); DNA binding / DNA-directed
DNA polymerase/ nucleic acid binding / nucleotide binding
/ zinc ion binding; K02324 DNA polymerase epsilon subunit
1 [EC:2.7.7.7]
Length=2145
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 38/67 (56%), Gaps = 1/67 (1%)
Query 14 VRKLRFEFPTWLLNFAVYRRFRNEMFLSFC-PERKSFLKEIKNEILFELDGPWKGMFLPA 72
++KL +P +LN V + N+ + + P RK++ + I FE+DGP+K M +PA
Sbjct 838 MKKLTISYPCVMLNVDVAKNNTNDQYQTLVDPVRKTYKSHSECSIEFEVDGPYKAMIIPA 897
Query 73 SEKSDSL 79
S++ L
Sbjct 898 SKEEGIL 904
> mmu:18973 Pole; polymerase (DNA directed), epsilon (EC:2.7.7.7);
K02324 DNA polymerase epsilon subunit 1 [EC:2.7.7.7]
Length=2283
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 32/61 (52%), Gaps = 1/61 (1%)
Query 16 KLRFEFPTWLLNFAVYRRFRNEMFLSFC-PERKSFLKEIKNEILFELDGPWKGMFLPASE 74
KL +P +LN V F N + P +++ +N I FE+DGP+ M LPAS+
Sbjct 887 KLTISYPGAMLNIMVKEGFTNHQYQELTEPSSLTYVTHSENSIFFEVDGPYLAMILPASK 946
Query 75 K 75
+
Sbjct 947 E 947
> ath:AT2G27120 TIL2; TIL2 (TILTED 2); DNA binding / DNA-directed
DNA polymerase/ nucleic acid binding / nucleotide binding
/ zinc ion binding; K02324 DNA polymerase epsilon subunit
1 [EC:2.7.7.7]
Length=2138
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 37/67 (55%), Gaps = 1/67 (1%)
Query 14 VRKLRFEFPTWLLNFAVYRRFRNEMFLSFC-PERKSFLKEIKNEILFELDGPWKGMFLPA 72
++K +P +LN V + N+ + + P RK++ + I FE+DGP+K M +PA
Sbjct 848 MKKFTISYPCVILNVDVAKNNSNDQYQTLVDPVRKTYNSRSECSIEFEVDGPYKAMIIPA 907
Query 73 SEKSDSL 79
S++ L
Sbjct 908 SKEEGIL 914
> hsa:5426 POLE, DKFZp434F222, FLJ21434, POLE1; polymerase (DNA
directed), epsilon (EC:2.7.7.7); K02324 DNA polymerase epsilon
subunit 1 [EC:2.7.7.7]
Length=2286
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 33/61 (54%), Gaps = 1/61 (1%)
Query 16 KLRFEFPTWLLNFAVYRRFRNEMFLSFC-PERKSFLKEIKNEILFELDGPWKGMFLPASE 74
K+ +P +LN V F N+ + P +++ +N I FE+DGP+ M LPAS+
Sbjct 887 KVTISYPGAMLNIMVKEGFTNDQYQELAEPSSLTYVTRSENSIFFEVDGPYLAMILPASK 946
Query 75 K 75
+
Sbjct 947 E 947
> cel:F33H2.5 hypothetical protein; K02324 DNA polymerase epsilon
subunit 1 [EC:2.7.7.7]
Length=2144
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 21/55 (38%), Positives = 33/55 (60%), Gaps = 1/55 (1%)
Query 21 FPTWLLNFAVYRRFRNEMFLSFCPERKSFLKEIKNEILFELDGPWKGMFLPASEK 75
+P +LN VY F N + + + S+ K +N I FE+DGP++ M LPAS++
Sbjct 872 YPGAMLNALVYEGFTNHQYHTL-EKDGSYSKSSENSIYFEVDGPYQCMILPASKE 925
> dre:553405 pole, si:dkeyp-98c11.2; polymerase (DNA directed),
epsilon (EC:2.7.7.7); K02324 DNA polymerase epsilon subunit
1 [EC:2.7.7.7]
Length=2284
Score = 37.7 bits (86), Expect = 0.010, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 30/61 (49%), Gaps = 1/61 (1%)
Query 16 KLRFEFPTWLLNFAVYRRFRNEMFLSFCPERK-SFLKEIKNEILFELDGPWKGMFLPASE 74
K+ +P +LN V F N + ++ +N I FE+DGP+ M LPAS+
Sbjct 886 KVTISYPGAMLNIMVKEGFTNHQYQELVDAASLTYETRAENSIFFEVDGPYLAMILPASK 945
Query 75 K 75
+
Sbjct 946 E 946
> pfa:PFF1595c VAR; erythrocyte membrane protein 1, PfEMP1; K13850
erythrocyte membrane protein 1
Length=2238
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 31 YRRFRNEMFLSFCPERKSFLKEIKNEILFELDG 63
Y R+ E +FC ERK LK+IK+E E +G
Sbjct 1218 YFRYLEEWGETFCRERKKRLKQIKHECKVEENG 1250
> pfa:PFD0625c VAR; erythrocyte membrane protein 1, PfEMP1; K13850
erythrocyte membrane protein 1
Length=2277
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 31 YRRFRNEMFLSFCPERKSFLKEIKNEILFELDGPWKG 67
Y R+ E +FC ERK L +I E + DGP G
Sbjct 1272 YFRYLEEWGQNFCKERKKRLDQIYRECKVDEDGPRDG 1308
> pfa:MAL7P1.212 VAR; erythrocyte membrane protein 1, PfEMP1;
K13850 erythrocyte membrane protein 1
Length=2269
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 25/49 (51%), Gaps = 3/49 (6%)
Query 22 PTWLLNFAV---YRRFRNEMFLSFCPERKSFLKEIKNEILFELDGPWKG 67
PT L++F Y R+ E +FC ERK LK+IK E + D G
Sbjct 1266 PTTLVDFISRPPYFRYLEEWGENFCKERKKRLKQIKVECKVDQDNDKNG 1314
> pfa:PFD0020c VAR; erythrocyte membrane protein 1, PfEMP1; K13850
erythrocyte membrane protein 1
Length=3467
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 23/45 (51%), Gaps = 3/45 (6%)
Query 22 PTWLLNFAV---YRRFRNEMFLSFCPERKSFLKEIKNEILFELDG 63
PT L NF Y R+ E +FC ERK L+E++ E E G
Sbjct 2441 PTTLTNFISRPPYFRYLEEWGETFCRERKKRLEEVRKECRGEYPG 2485
> pfa:PF11_0159 conserved Plasmodium protein
Length=145
Score = 28.9 bits (63), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 16/59 (27%), Positives = 33/59 (55%), Gaps = 3/59 (5%)
Query 18 RFEFPTWLLNFAVYRRFRNEMFLSFCPERKSFLKEIKN-EILFELDGPWKGMFLPASEK 75
+++FP W +N+ + ++ +N +F++ + F K I++ LF WKG + P+ K
Sbjct 34 QYKFPKWKINYDIVKKNQNHIFINRVND--YFQKLIESLSRLFWKGRKWKGTYWPSKWK 90
> pfa:PF07_0048 VAR; erythrocyte membrane protein 1, PfEMP1; K13850
erythrocyte membrane protein 1
Length=2215
Score = 28.5 bits (62), Expect = 5.9, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 3/40 (7%)
Query 22 PTWLLNFAV---YRRFRNEMFLSFCPERKSFLKEIKNEIL 58
PT L +F Y R+ E +FC ERK L +IK+E +
Sbjct 1208 PTTLTDFISRPPYFRYLEEWGQNFCKERKKRLAQIKHECM 1247
> pfa:PFD0615c VAR; erythrocyte membrane protein 1, PfEMP1; K13850
erythrocyte membrane protein 1
Length=2209
Score = 27.7 bits (60), Expect = 9.0, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 27/55 (49%), Gaps = 4/55 (7%)
Query 2 ISSSSSSSSSRGVRKLRFEFPTWLLNFAVYRRFRNEMFLSFCPERKSFLKEIKNE 56
+ S+ S ++RG + F + Y R+ E +FC ERK LK+IK E
Sbjct 1188 LQSTDSKDAARGEKTPLDSF----IKRPPYFRYLEEWGQNFCKERKKRLKDIKYE 1238
Lambda K H
0.322 0.135 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2063098576
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40