bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5271_orf3
Length=109
Score E
Sequences producing significant alignments: (Bits) Value
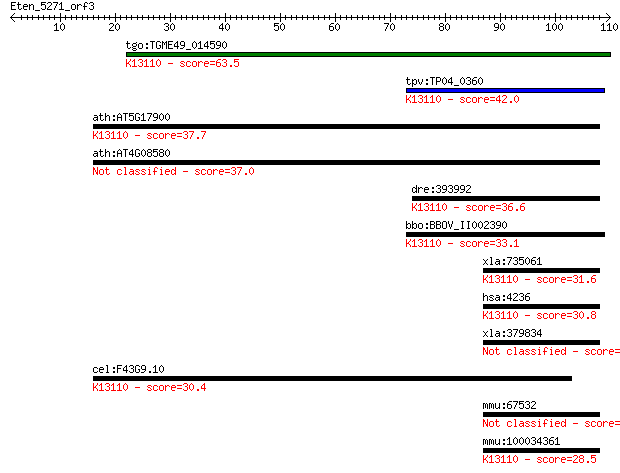
tgo:TGME49_014590 microfibrillar-associated protein 1, putativ... 63.5 2e-10
tpv:TP04_0360 hypothetical protein; K13110 microfibrillar-asso... 42.0 5e-04
ath:AT5G17900 hypothetical protein; K13110 microfibrillar-asso... 37.7 0.010
ath:AT4G08580 hypothetical protein 37.0 0.017
dre:393992 mfap1, MGC56551, zgc:56551; microfibrillar-associat... 36.6 0.019
bbo:BBOV_II002390 18.m06195; micro-fibrillar-associated protei... 33.1 0.25
xla:735061 hypothetical protein MGC85054; K13110 microfibrilla... 31.6 0.71
hsa:4236 MFAP1; microfibrillar-associated protein 1; K13110 mi... 30.8 1.1
xla:379834 mfap1, MGC53463; microfibrillar-associated protein 1 30.4
cel:F43G9.10 hypothetical protein; K13110 microfibrillar-assoc... 30.4 1.6
mmu:67532 Mfap1a, 4432409M24Rik, Mfap1; microfibrillar-associa... 28.5 5.2
mmu:100034361 Mfap1b; microfibrillar-associated protein 1B; K1... 28.5 5.2
> tgo:TGME49_014590 microfibrillar-associated protein 1, putative
; K13110 microfibrillar-associated protein 1
Length=438
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 39/94 (41%), Positives = 53/94 (56%), Gaps = 26/94 (27%)
Query 22 RKLETKQMVYAVLAREDEEAADAPVIPVEAQQQQQQQQQQQKNLMGSE------EMPDDT 75
RK E+K+++Y L +EDEE + N++G + E+PDDT
Sbjct 222 RKRESKKLLYEALQKEDEE--------------------MRTNMLGEQLQEEDCELPDDT 261
Query 76 DGLDAAQEYEDWKLRELERIRRDKEEQLEREKFL 109
DGLDA EYE WK REL RI+RD+EE+ R+ FL
Sbjct 262 DGLDAEAEYEAWKARELLRIKRDQEERQARDNFL 295
> tpv:TP04_0360 hypothetical protein; K13110 microfibrillar-associated
protein 1
Length=408
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 24/36 (66%), Positives = 27/36 (75%), Gaps = 4/36 (11%)
Query 73 DDTDGLDAAQEYEDWKLRELERIRRDKEEQLEREKF 108
DDTD D +EYE WK+REL+RI RDKE EREKF
Sbjct 244 DDTDTFDE-KEYELWKIRELKRILRDKE---EREKF 275
> ath:AT5G17900 hypothetical protein; K13110 microfibrillar-associated
protein 1
Length=435
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 35/97 (36%), Positives = 51/97 (52%), Gaps = 26/97 (26%)
Query 16 QQQVEQRKLETKQMVYAVLAREDEEAADAPVIPVEAQQQQQQQQQQQKNLMGSEEMPDDT 75
++++EQRKLETKQ+V + R+DEE +KN++ E D
Sbjct 212 KRKLEQRKLETKQIVVEEV-RKDEEI--------------------RKNILLEEANIGDV 250
Query 76 ---DGLDAAQEYEDWKLRELERIR--RDKEEQLEREK 107
D L+ A+EYE WK RE+ RI+ RD E + RE+
Sbjct 251 ETDDELNEAEEYEVWKTREIGRIKRERDAREAMLRER 287
> ath:AT4G08580 hypothetical protein
Length=435
Score = 37.0 bits (84), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 34/97 (35%), Positives = 51/97 (52%), Gaps = 26/97 (26%)
Query 16 QQQVEQRKLETKQMVYAVLAREDEEAADAPVIPVEAQQQQQQQQQQQKNLMGSEEMPDDT 75
++++EQRK+ETKQ+V + R+DEE +KN++ E D
Sbjct 212 KRKLEQRKIETKQIVVEEV-RKDEEI--------------------RKNILLEEANIGDV 250
Query 76 ---DGLDAAQEYEDWKLRELERIR--RDKEEQLEREK 107
D L+ A+EYE WK RE+ RI+ RD E + RE+
Sbjct 251 ETDDELNEAEEYEVWKTREIGRIKRERDAREAMLRER 287
> dre:393992 mfap1, MGC56551, zgc:56551; microfibrillar-associated
protein 1; K13110 microfibrillar-associated protein 1
Length=437
Score = 36.6 bits (83), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 22/36 (61%), Positives = 29/36 (80%), Gaps = 2/36 (5%)
Query 74 DTDGLDAAQEYEDWKLRELERIRRDKE--EQLEREK 107
DTDG + +EYE WK+REL+RI+RD+E E LE+EK
Sbjct 265 DTDGENEEEEYEAWKVRELKRIKRDRESREALEKEK 300
> bbo:BBOV_II002390 18.m06195; micro-fibrillar-associated protein
1 C-terminus containing protein; K13110 microfibrillar-associated
protein 1
Length=437
Score = 33.1 bits (74), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 18/36 (50%), Positives = 24/36 (66%), Gaps = 1/36 (2%)
Query 73 DDTDGLDAAQEYEDWKLRELERIRRDKEEQLEREKF 108
DD D L +EYE WK+REL+RI RD+ E+ E+
Sbjct 264 DDKDEL-TEEEYELWKIRELKRIIRDRNERNAHERL 298
> xla:735061 hypothetical protein MGC85054; K13110 microfibrillar-associated
protein 1
Length=442
Score = 31.6 bits (70), Expect = 0.71, Method: Compositional matrix adjust.
Identities = 15/23 (65%), Positives = 21/23 (91%), Gaps = 2/23 (8%)
Query 87 WKLRELERIRRDKEEQ--LEREK 107
WK+REL+RI+RD+EE+ LE+EK
Sbjct 282 WKVRELKRIKRDREEREALEKEK 304
> hsa:4236 MFAP1; microfibrillar-associated protein 1; K13110
microfibrillar-associated protein 1
Length=439
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 15/23 (65%), Positives = 20/23 (86%), Gaps = 2/23 (8%)
Query 87 WKLRELERIRRDKE--EQLEREK 107
WK+REL+RI+RD+E E LE+EK
Sbjct 279 WKVRELKRIKRDREDREALEKEK 301
> xla:379834 mfap1, MGC53463; microfibrillar-associated protein
1
Length=441
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 14/23 (60%), Positives = 21/23 (91%), Gaps = 2/23 (8%)
Query 87 WKLRELERIRRDKEEQ--LEREK 107
WK+REL+RI+RD+EE+ +E+EK
Sbjct 281 WKVRELKRIKRDREEREAMEKEK 303
> cel:F43G9.10 hypothetical protein; K13110 microfibrillar-associated
protein 1
Length=466
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 24/87 (27%), Positives = 51/87 (58%), Gaps = 19/87 (21%)
Query 16 QQQVEQRKLETKQMVYAVLAREDEEAADAPVIPVEAQQQQQQQQQQQKNLMGSEEMPDDT 75
+++ E+RK E+ ++V VL ++EEAA ++++ + + + S D+T
Sbjct 255 EKRAEERKRESAKLVEKVL--QEEEAA-------------EKRKTEDRVDLSSVLTDDET 299
Query 76 DGLDAAQEYEDWKLRELERIRRDKEEQ 102
+ + YE WKLRE++R++R+++E+
Sbjct 300 ENM----AYEAWKLREMKRLKRNRDER 322
> mmu:67532 Mfap1a, 4432409M24Rik, Mfap1; microfibrillar-associated
protein 1A
Length=439
Score = 28.5 bits (62), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 14/23 (60%), Positives = 20/23 (86%), Gaps = 2/23 (8%)
Query 87 WKLRELERIRRDKE--EQLEREK 107
WK+REL+RI+R++E E LE+EK
Sbjct 279 WKVRELKRIKREREDREALEKEK 301
> mmu:100034361 Mfap1b; microfibrillar-associated protein 1B;
K13110 microfibrillar-associated protein 1
Length=439
Score = 28.5 bits (62), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 14/23 (60%), Positives = 20/23 (86%), Gaps = 2/23 (8%)
Query 87 WKLRELERIRRDKE--EQLEREK 107
WK+REL+RI+R++E E LE+EK
Sbjct 279 WKVRELKRIKREREDREALEKEK 301
Lambda K H
0.307 0.122 0.317
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2072286120
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40