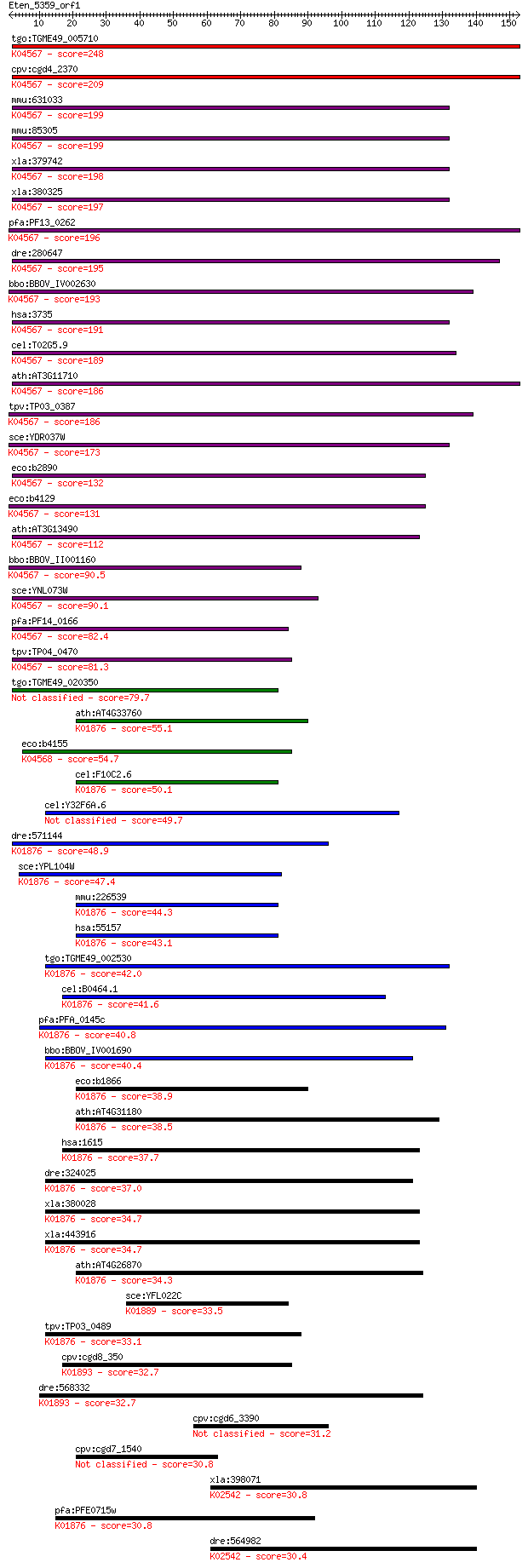
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5359_orf1
Length=152
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_005710 lysyl-tRNA synthetase, putative (EC:6.1.1.6)... 248 4e-66
cpv:cgd4_2370 lysyl-tRNA synthetase (NOB+tRNA synthetase) ; K0... 209 4e-54
mmu:631033 lysyl-tRNA synthetase-like; K04567 lysyl-tRNA synth... 199 3e-51
mmu:85305 Kars, AA589550, AL024334, AL033315, AL033367, D8Ertd... 199 4e-51
xla:379742 kars, MGC53345; lysyl-tRNA synthetase (EC:6.1.1.6);... 198 7e-51
xla:380325 kars, MGC52992, kars2, krs, krs-1; lysyl-tRNA synth... 197 1e-50
pfa:PF13_0262 lysine-tRNA ligase (EC:6.1.1.6); K04567 lysyl-tR... 196 2e-50
dre:280647 kars, cb530, zgc:92483; lysyl-tRNA synthetase (EC:6... 195 6e-50
bbo:BBOV_IV002630 21.m02971; lysyl-tRNA synthetase (EC:6.1.1.6... 193 2e-49
hsa:3735 KARS, CMTRIB, KARS2, KIAA0070, KRS; lysyl-tRNA synthe... 191 6e-49
cel:T02G5.9 krs-1; lysyl (K) tRNA Synthetase family member (kr... 189 2e-48
ath:AT3G11710 ATKRS-1 (ARABIDOPSIS THALIANA LYSYL-TRNA SYNTHET... 186 3e-47
tpv:TP03_0387 lysyl-tRNA synthetase (EC:6.1.1.6); K04567 lysyl... 186 3e-47
sce:YDR037W KRS1, GCD5; Krs1p (EC:6.1.1.6); K04567 lysyl-tRNA ... 173 2e-43
eco:b2890 lysS, asuD, ECK2885, herC, JW2858; lysine tRNA synth... 132 5e-31
eco:b4129 lysU, ECK4123, JW4090; lysine tRNA synthetase, induc... 131 1e-30
ath:AT3G13490 OVA5; OVA5 (OVULE ABORTION 5); ATP binding / ami... 112 6e-25
bbo:BBOV_II001160 18.m06086; lysyl-tRNA synthetase; K04567 lys... 90.5 2e-18
sce:YNL073W MSK1; Mitochondrial lysine-tRNA synthetase, requir... 90.1 2e-18
pfa:PF14_0166 lysine-tRNA ligase, putative (EC:6.1.1.6); K0456... 82.4 5e-16
tpv:TP04_0470 lysyl-tRNA synthetase; K04567 lysyl-tRNA synthet... 81.3 1e-15
tgo:TGME49_020350 lysyl-tRNA synthetase, putative (EC:6.1.1.12... 79.7 3e-15
ath:AT4G33760 tRNA synthetase class II (D, K and N) family pro... 55.1 1e-07
eco:b4155 yjeA, ECK4151, genX, JW4116, poxA; EF-P-lysine34-lys... 54.7 1e-07
cel:F10C2.6 drs-2; aspartyl(D) tRNA Synthetase family member (... 50.1 3e-06
cel:Y32F6A.6 hypothetical protein 49.7 4e-06
dre:571144 dars2, si:dkey-21n10.1; aspartyl-tRNA synthetase 2,... 48.9 7e-06
sce:YPL104W MSD1, LPG5; Msd1p (EC:6.1.1.12); K01876 aspartyl-t... 47.4 2e-05
mmu:226539 Dars2, 5830468K18Rik, MGC99963; aspartyl-tRNA synth... 44.3 1e-04
hsa:55157 DARS2, ASPRS, FLJ10514, LBSL, MT-ASPRS, RP3-383J4.2;... 43.1 3e-04
tgo:TGME49_002530 aspartyl-tRNA synthetase, putative (EC:6.1.1... 42.0 8e-04
cel:B0464.1 drs-1; aspartyl(D) tRNA Synthetase family member (... 41.6 0.001
pfa:PFA_0145c aspartyl-tRNA synthetase, putative (EC:6.1.1.12)... 40.8 0.002
bbo:BBOV_IV001690 21.m02784; aspartyl-tRNA synthetase (EC:6.1.... 40.4 0.002
eco:b1866 aspS, ECK1867, JW1855, tls; aspartyl-tRNA synthetase... 38.9 0.006
ath:AT4G31180 aspartyl-tRNA synthetase, putative / aspartate--... 38.5 0.009
hsa:1615 DARS, DKFZp781B11202, MGC111579; aspartyl-tRNA synthe... 37.7 0.013
dre:324025 dars, MGC154056, wu:fc17a11, zgc:154056; aspartyl-t... 37.0 0.023
xla:380028 dars, MGC53970; aspartyl-tRNA synthetase (EC:6.1.1.... 34.7 0.11
xla:443916 dars, MGC80207; aspartyl-tRNA synthetase (EC:6.1.1.... 34.7 0.12
ath:AT4G26870 aspartyl-tRNA synthetase, putative / aspartate--... 34.3 0.14
sce:YFL022C FRS2; Frs2p (EC:6.1.1.20); K01889 phenylalanyl-tRN... 33.5 0.25
tpv:TP03_0489 aspartyl-tRNA synthetase (EC:6.1.1.12); K01876 a... 33.1 0.37
cpv:cgd8_350 asparaginyl-tRNA synthetase (NOB+tRNA synthase) ;... 32.7 0.41
dre:568332 nars, fb57a10, si:dkey-274m14.2, wu:fb57a10; aspara... 32.7 0.44
cpv:cgd6_3390 membrane associated protein with 2 transmembrane... 31.2 1.3
cpv:cgd7_1540 aspartate--tRNA ligase 30.8 1.7
xla:398071 mcm6, MGC85070, mis5, mmcm6; minichromosome mainten... 30.8 1.7
pfa:PFE0715w asparagine-tRNA ligase, putative (EC:6.1.1.12); K... 30.8 1.8
dre:564982 mcm6l, MGC112244, zgc:112244; MCM6 minichromosome m... 30.4 2.5
> tgo:TGME49_005710 lysyl-tRNA synthetase, putative (EC:6.1.1.6);
K04567 lysyl-tRNA synthetase, class II [EC:6.1.1.6]
Length=658
Score = 248 bits (634), Expect = 4e-66, Method: Compositional matrix adjust.
Identities = 111/151 (73%), Positives = 132/151 (87%), Gaps = 0/151 (0%)
Query 2 AKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACE 61
A+PF THHNDL+ DLYMR+APEL+LKML VGGMD+VFEIGKNFRNEGIDMTHNPEFTACE
Sbjct 339 ARPFKTHHNDLDMDLYMRIAPELFLKMLIVGGMDRVFEIGKNFRNEGIDMTHNPEFTACE 398
Query 62 FYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVEE 121
FYWAYADYND M TE+++S MVM++HG+Y+I FHP+GP V LDFTPP+ RLS+VEE
Sbjct 399 FYWAYADYNDLMNFTEELVSSMVMALHGTYQIQFHPDGPGSEPVTLDFTPPFARLSMVEE 458
Query 122 IEKQAKVTLPRPLDGPECLACLKDLMEKHNI 152
IEKQA VTLPRPLDG EC+A +K+L+ K+ +
Sbjct 459 IEKQAGVTLPRPLDGDECIAFMKELLIKNKV 489
> cpv:cgd4_2370 lysyl-tRNA synthetase (NOB+tRNA synthetase) ;
K04567 lysyl-tRNA synthetase, class II [EC:6.1.1.6]
Length=559
Score = 209 bits (531), Expect = 4e-54, Method: Compositional matrix adjust.
Identities = 96/151 (63%), Positives = 118/151 (78%), Gaps = 0/151 (0%)
Query 2 AKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACE 61
A+PF+T+HN+L T LYMR+APELYLK L VGG+DKV+EIGKNFRNEGID+THNPEFTA E
Sbjct 252 ARPFITYHNELETQLYMRIAPELYLKQLIVGGLDKVYEIGKNFRNEGIDLTHNPEFTAME 311
Query 62 FYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVEE 121
FY AYADY D M LTE++IS +V+ IHGS KIP+HP+GP G +E+DFT P++R S VEE
Sbjct 312 FYMAYADYYDLMDLTEELISGLVLEIHGSLKIPYHPDGPEGKCIEIDFTTPWKRFSFVEE 371
Query 122 IEKQAKVTLPRPLDGPECLACLKDLMEKHNI 152
IE L RPLD E + + ++ EKH I
Sbjct 372 IESGLGEKLKRPLDSQENIDFMVEMCEKHEI 402
> mmu:631033 lysyl-tRNA synthetase-like; K04567 lysyl-tRNA synthetase,
class II [EC:6.1.1.6]
Length=595
Score = 199 bits (505), Expect = 3e-51, Method: Compositional matrix adjust.
Identities = 88/130 (67%), Positives = 109/130 (83%), Gaps = 0/130 (0%)
Query 2 AKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACE 61
AKPF+T+HN+L+ +LYMR+APELY KML VGG+D+V+EIG+ FRNEGID+THNPEFT CE
Sbjct 278 AKPFITYHNELDMNLYMRIAPELYHKMLVVGGIDRVYEIGRQFRNEGIDLTHNPEFTTCE 337
Query 62 FYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVEE 121
FY AYADY+D M +TE M+S MV SI GSYKI +HP+GP G E+DFTPP+RR+S+VEE
Sbjct 338 FYMAYADYHDLMEITEKMLSGMVKSITGSYKITYHPDGPEGQAYEVDFTPPFRRISMVEE 397
Query 122 IEKQAKVTLP 131
+EK V LP
Sbjct 398 LEKALGVKLP 407
> mmu:85305 Kars, AA589550, AL024334, AL033315, AL033367, D8Ertd698e,
D8Wsu108e, LysRS, mKIAA0070; lysyl-tRNA synthetase (EC:6.1.1.6);
K04567 lysyl-tRNA synthetase, class II [EC:6.1.1.6]
Length=624
Score = 199 bits (505), Expect = 4e-51, Method: Compositional matrix adjust.
Identities = 88/130 (67%), Positives = 109/130 (83%), Gaps = 0/130 (0%)
Query 2 AKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACE 61
AKPF+T+HN+L+ +LYMR+APELY KML VGG+D+V+EIG+ FRNEGID+THNPEFT CE
Sbjct 307 AKPFITYHNELDMNLYMRIAPELYHKMLVVGGIDRVYEIGRQFRNEGIDLTHNPEFTTCE 366
Query 62 FYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVEE 121
FY AYADY+D M +TE M+S MV SI GSYKI +HP+GP G E+DFTPP+RR+S+VEE
Sbjct 367 FYMAYADYHDLMEITEKMLSGMVKSITGSYKITYHPDGPEGQAYEVDFTPPFRRISMVEE 426
Query 122 IEKQAKVTLP 131
+EK V LP
Sbjct 427 LEKALGVKLP 436
> xla:379742 kars, MGC53345; lysyl-tRNA synthetase (EC:6.1.1.6);
K04567 lysyl-tRNA synthetase, class II [EC:6.1.1.6]
Length=605
Score = 198 bits (503), Expect = 7e-51, Method: Compositional matrix adjust.
Identities = 87/130 (66%), Positives = 106/130 (81%), Gaps = 0/130 (0%)
Query 2 AKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACE 61
AKPF+THHN+L+ LYMR+APELY KML VGG+D+V+EIG+ FRNEGID+THNPEFT CE
Sbjct 292 AKPFITHHNELDMKLYMRIAPELYHKMLVVGGIDRVYEIGRQFRNEGIDLTHNPEFTTCE 351
Query 62 FYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVEE 121
FY AYADYND M +TE ++S MV I G YK+ +HPEGP GPV E+DFTPP+RR+S+V E
Sbjct 352 FYMAYADYNDLMEITEKLLSGMVKHITGGYKVTYHPEGPEGPVYEIDFTPPFRRISMVHE 411
Query 122 IEKQAKVTLP 131
+EK LP
Sbjct 412 LEKALGKKLP 421
> xla:380325 kars, MGC52992, kars2, krs, krs-1; lysyl-tRNA synthetase
(EC:6.1.1.6); K04567 lysyl-tRNA synthetase, class II
[EC:6.1.1.6]
Length=605
Score = 197 bits (500), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 87/130 (66%), Positives = 106/130 (81%), Gaps = 0/130 (0%)
Query 2 AKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACE 61
AKPF+T+HNDL+ LYMR+APELY KML VGG+D+V+EIG+ FRNEGID+THNPEFT CE
Sbjct 292 AKPFITYHNDLDMKLYMRIAPELYHKMLVVGGIDRVYEIGRQFRNEGIDLTHNPEFTTCE 351
Query 62 FYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVEE 121
FY AYADYND M +TE ++S MV I G YK+P+HPEGP GP E+DFTPP+RR+S+V E
Sbjct 352 FYMAYADYNDLMEITEKLLSGMVKHITGGYKVPYHPEGPEGPEYEIDFTPPFRRISMVHE 411
Query 122 IEKQAKVTLP 131
+EK LP
Sbjct 412 LEKALGKKLP 421
> pfa:PF13_0262 lysine-tRNA ligase (EC:6.1.1.6); K04567 lysyl-tRNA
synthetase, class II [EC:6.1.1.6]
Length=583
Score = 196 bits (499), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 87/152 (57%), Positives = 118/152 (77%), Gaps = 0/152 (0%)
Query 1 NAKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTAC 60
NA+PF+THHNDL+ DLY+R+A EL LKML VGG+DKV+EIGK FRNEGID THNPEFT+C
Sbjct 286 NARPFITHHNDLDLDLYLRIATELPLKMLIVGGIDKVYEIGKVFRNEGIDNTHNPEFTSC 345
Query 61 EFYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVE 120
EFYWAYADYND ++ +ED S++V + G+YKI ++ +GP +E+DFTPPY ++S+VE
Sbjct 346 EFYWAYADYNDLIKWSEDFFSQLVYHLFGTYKISYNKDGPENQPIEIDFTPPYPKVSIVE 405
Query 121 EIEKQAKVTLPRPLDGPECLACLKDLMEKHNI 152
EIEK L +P D E + + +++++H I
Sbjct 406 EIEKVTNTILEQPFDSNETIEKMINIIKEHKI 437
> dre:280647 kars, cb530, zgc:92483; lysyl-tRNA synthetase (EC:6.1.1.6);
K04567 lysyl-tRNA synthetase, class II [EC:6.1.1.6]
Length=602
Score = 195 bits (495), Expect = 6e-50, Method: Compositional matrix adjust.
Identities = 89/147 (60%), Positives = 113/147 (76%), Gaps = 2/147 (1%)
Query 2 AKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACE 61
AKPF+T+HNDLN +LYMR+APELY KML VGG+D+V+EIG+ FRNEGID+THNPEFT CE
Sbjct 293 AKPFITYHNDLNMNLYMRIAPELYHKMLVVGGIDRVYEIGRQFRNEGIDLTHNPEFTTCE 352
Query 62 FYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVEE 121
FY AYADY+D M +TE ++S MV I G+YK+ +HP+GP G E+DFTPP+RR+S+ +E
Sbjct 353 FYMAYADYHDLMEITEKLLSGMVKHITGAYKVTYHPDGPEGQAYEIDFTPPFRRISMTQE 412
Query 122 IEKQAKVTLPRP--LDGPECLACLKDL 146
+EK+ V P P D E L DL
Sbjct 413 LEKELGVKFPPPDTYDSDEMRKFLDDL 439
> bbo:BBOV_IV002630 21.m02971; lysyl-tRNA synthetase (EC:6.1.1.6);
K04567 lysyl-tRNA synthetase, class II [EC:6.1.1.6]
Length=548
Score = 193 bits (490), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 82/138 (59%), Positives = 111/138 (80%), Gaps = 0/138 (0%)
Query 1 NAKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTAC 60
+A+PF++HHNDL+ DL++RVAPEL LKM+ VGG +KVFEIGK FRNEGIDMTHNPEFT+C
Sbjct 214 SARPFISHHNDLDIDLFLRVAPELPLKMIVVGGFEKVFEIGKCFRNEGIDMTHNPEFTSC 273
Query 61 EFYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVE 120
EFYWAYADYND ++LTE+ +S +V+ +HG Y +P+HP+GP GP + LDF PPY R+SL++
Sbjct 274 EFYWAYADYNDLIQLTEEYLSSLVLELHGGYTLPYHPDGPEGPEIILDFKPPYDRISLID 333
Query 121 EIEKQAKVTLPRPLDGPE 138
+++ + P + E
Sbjct 334 ALKQGTGIDFVPPYNSDE 351
> hsa:3735 KARS, CMTRIB, KARS2, KIAA0070, KRS; lysyl-tRNA synthetase
(EC:6.1.1.6); K04567 lysyl-tRNA synthetase, class II
[EC:6.1.1.6]
Length=625
Score = 191 bits (486), Expect = 6e-49, Method: Compositional matrix adjust.
Identities = 83/130 (63%), Positives = 108/130 (83%), Gaps = 0/130 (0%)
Query 2 AKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACE 61
AKPF+T+HN+L+ +LYMR+APELY KML VGG+D+V+EIG+ FRNEGID+THNPEFT CE
Sbjct 308 AKPFITYHNELDMNLYMRIAPELYHKMLVVGGIDRVYEIGRQFRNEGIDLTHNPEFTTCE 367
Query 62 FYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVEE 121
FY AYADY+D M +TE M+S MV I GSYK+ +HP+GP G ++DFTPP+RR+++VEE
Sbjct 368 FYMAYADYHDLMEITEKMVSGMVKHITGSYKVTYHPDGPEGQAYDVDFTPPFRRINMVEE 427
Query 122 IEKQAKVTLP 131
+EK + LP
Sbjct 428 LEKALGMKLP 437
> cel:T02G5.9 krs-1; lysyl (K) tRNA Synthetase family member (krs-1);
K04567 lysyl-tRNA synthetase, class II [EC:6.1.1.6]
Length=572
Score = 189 bits (481), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 84/133 (63%), Positives = 110/133 (82%), Gaps = 1/133 (0%)
Query 2 AKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACE 61
AKPF+THHNDL+ +L++RVAPELY KML VGG+D+V+E+G+ FRNEGID+THNPEFT CE
Sbjct 259 AKPFITHHNDLDMNLFLRVAPELYHKMLVVGGIDRVYEVGRLFRNEGIDLTHNPEFTTCE 318
Query 62 FYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNG-PVVELDFTPPYRRLSLVE 120
FY AYADY D ++LTED++S MVMSI G+YKI +HP GPN PV E+DFTPP++R+ + +
Sbjct 319 FYMAYADYEDVIQLTEDLLSSMVMSIKGTYKIEYHPNGPNTEPVYEVDFTPPFKRVHMYD 378
Query 121 EIEKQAKVTLPRP 133
+ ++ TLP P
Sbjct 379 GLAEKLGATLPDP 391
> ath:AT3G11710 ATKRS-1 (ARABIDOPSIS THALIANA LYSYL-TRNA SYNTHETASE
1); ATP binding / aminoacyl-tRNA ligase/ lysine-tRNA ligase/
nucleic acid binding / nucleotide binding (EC:6.1.1.6);
K04567 lysyl-tRNA synthetase, class II [EC:6.1.1.6]
Length=626
Score = 186 bits (472), Expect = 3e-47, Method: Compositional matrix adjust.
Identities = 84/151 (55%), Positives = 112/151 (74%), Gaps = 0/151 (0%)
Query 2 AKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACE 61
A+PFVTHHNDL+ LYMR+APELYLK L VGG+++V+EIGK FRNEGID+THNPEFT CE
Sbjct 316 ARPFVTHHNDLDMRLYMRIAPELYLKQLIVGGLERVYEIGKQFRNEGIDLTHNPEFTTCE 375
Query 62 FYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVEE 121
FY A+ADYND M +TE M+S MV + G YKI ++ G + +E+DFTPP+RR+ ++ E
Sbjct 376 FYMAFADYNDLMEMTEVMLSGMVKELTGGYKIKYNANGYDKDPIEIDFTPPFRRIEMIGE 435
Query 122 IEKQAKVTLPRPLDGPECLACLKDLMEKHNI 152
+EK AK+ +P+ L E L D + ++
Sbjct 436 LEKVAKLNIPKDLASEEANKYLIDACARFDV 466
> tpv:TP03_0387 lysyl-tRNA synthetase (EC:6.1.1.6); K04567 lysyl-tRNA
synthetase, class II [EC:6.1.1.6]
Length=551
Score = 186 bits (471), Expect = 3e-47, Method: Compositional matrix adjust.
Identities = 81/138 (58%), Positives = 111/138 (80%), Gaps = 0/138 (0%)
Query 1 NAKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTAC 60
+AKPF+THHN+L+ DL+MR+APEL LK++ +GG +KVFEIGK FRNEGID THNPEFT+C
Sbjct 250 SAKPFITHHNELDLDLFMRIAPELPLKLIIIGGFEKVFEIGKCFRNEGIDPTHNPEFTSC 309
Query 61 EFYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVE 120
EFYWAYADY+D M+LTE+ +S +V + G Y++ +HP+GP+ V +DFTPP+ ++S+VE
Sbjct 310 EFYWAYADYHDLMKLTEEFLSSLVFELFGKYEVLYHPDGPDTEGVVIDFTPPFNKVSMVE 369
Query 121 EIEKQAKVTLPRPLDGPE 138
E+E + K+ L P D E
Sbjct 370 ELENKMKMKLSPPYDSQE 387
> sce:YDR037W KRS1, GCD5; Krs1p (EC:6.1.1.6); K04567 lysyl-tRNA
synthetase, class II [EC:6.1.1.6]
Length=591
Score = 173 bits (439), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 81/133 (60%), Positives = 103/133 (77%), Gaps = 3/133 (2%)
Query 1 NAKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTAC 60
AKPF+THHNDL+ D+YMR+APEL+LK L VGG+D+V+EIG+ FRNEGIDMTHNPEFT C
Sbjct 282 TAKPFITHHNDLDMDMYMRIAPELFLKQLVVGGLDRVYEIGRQFRNEGIDMTHNPEFTTC 341
Query 61 EFYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPV--VELDFTPPYRRLSL 118
EFY AYAD D M +TE M SEMV I GSY I +HP+ P P +EL+F+ P++R+++
Sbjct 342 EFYQAYADVYDLMDMTELMFSEMVKEITGSYIIKYHPD-PADPAKELELNFSRPWKRINM 400
Query 119 VEEIEKQAKVTLP 131
+EE+EK V P
Sbjct 401 IEELEKVFNVKFP 413
> eco:b2890 lysS, asuD, ECK2885, herC, JW2858; lysine tRNA synthetase,
constitutive (EC:6.1.1.6); K04567 lysyl-tRNA synthetase,
class II [EC:6.1.1.6]
Length=505
Score = 132 bits (332), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 61/123 (49%), Positives = 82/123 (66%), Gaps = 7/123 (5%)
Query 2 AKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACE 61
A+PF+THHN L+ D+Y+R+APELYLK L VGG ++VFEI +NFRNEGI + HNPEFT E
Sbjct 220 ARPFITHHNALDLDMYLRIAPELYLKRLVVGGFERVFEINRNFRNEGISVRHNPEFTMME 279
Query 62 FYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVEE 121
Y AYADY D + LTE + + I G ++ + V LDF P+ +L++ E
Sbjct 280 LYMAYADYKDLIELTESLFRTLAQDILGKTEVTYGD-------VTLDFGKPFEKLTMREA 332
Query 122 IEK 124
I+K
Sbjct 333 IKK 335
> eco:b4129 lysU, ECK4123, JW4090; lysine tRNA synthetase, inducible
(EC:6.1.1.6); K04567 lysyl-tRNA synthetase, class II
[EC:6.1.1.6]
Length=505
Score = 131 bits (329), Expect = 1e-30, Method: Composition-based stats.
Identities = 61/124 (49%), Positives = 85/124 (68%), Gaps = 7/124 (5%)
Query 1 NAKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTAC 60
+A+PF+THHN L+ D+Y+R+APELYLK L VGG ++VFEI +NFRNEGI + HNPEFT
Sbjct 219 SARPFITHHNALDLDMYLRIAPELYLKRLVVGGFERVFEINRNFRNEGISVRHNPEFTMM 278
Query 61 EFYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVE 120
E Y AYADY+D + LTE + + + G+ K+ + G V DF P+ +L++ E
Sbjct 279 ELYMAYADYHDLIELTESLFRTLAQEVLGTTKVTY------GEHV-FDFGKPFEKLTMRE 331
Query 121 EIEK 124
I+K
Sbjct 332 AIKK 335
> ath:AT3G13490 OVA5; OVA5 (OVULE ABORTION 5); ATP binding / aminoacyl-tRNA
ligase/ lysine-tRNA ligase/ nucleic acid binding
/ nucleotide binding (EC:6.1.1.6); K04567 lysyl-tRNA synthetase,
class II [EC:6.1.1.6]
Length=602
Score = 112 bits (279), Expect = 6e-25, Method: Composition-based stats.
Identities = 56/124 (45%), Positives = 79/124 (63%), Gaps = 10/124 (8%)
Query 2 AKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACE 61
A+PFVT HN L DLY+R+A EL+LK + VGG +KV+EIG+ FRNEGI HNPEFT E
Sbjct 288 ARPFVTFHNSLGRDLYLRIATELHLKRMLVGGFEKVYEIGRIFRNEGISTRHNPEFTTIE 347
Query 62 FYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLS---L 118
Y AY+DY+ M + E ++++ M+++G I + E+ P+RR + L
Sbjct 348 MYEAYSDYHSMMDMAELIVTQCSMAVNGKLTIDYQG-------TEICLERPWRRETMHNL 400
Query 119 VEEI 122
V+EI
Sbjct 401 VKEI 404
> bbo:BBOV_II001160 18.m06086; lysyl-tRNA synthetase; K04567 lysyl-tRNA
synthetase, class II [EC:6.1.1.6]
Length=522
Score = 90.5 bits (223), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 43/88 (48%), Positives = 57/88 (64%), Gaps = 1/88 (1%)
Query 1 NAKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGM-DKVFEIGKNFRNEGIDMTHNPEFTA 59
NA+PF T L+ +LY+R+APE +LK L VGG+ DK+FEIGK FRNEG H+PEFT
Sbjct 255 NAQPFKTRSEALDENLYLRIAPEFFLKRLIVGGLGDKIFEIGKCFRNEGTSARHSPEFTM 314
Query 60 CEFYWAYADYNDTMRLTEDMISEMVMSI 87
E Y A+ D + L E++I + I
Sbjct 315 IEIYQQMANAGDMINLLEEIIETLAKKI 342
> sce:YNL073W MSK1; Mitochondrial lysine-tRNA synthetase, required
for import of both aminoacylated and deacylated forms of
tRNA(Lys) into mitochondria and for aminoacylation of mitochondrially
encoded tRNA(Lys) (EC:6.1.1.6); K04567 lysyl-tRNA
synthetase, class II [EC:6.1.1.6]
Length=576
Score = 90.1 bits (222), Expect = 2e-18, Method: Composition-based stats.
Identities = 42/91 (46%), Positives = 60/91 (65%), Gaps = 1/91 (1%)
Query 2 AKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACE 61
AKPF+T D + L +R+APEL+LK L + G+ KV+EIGK FRNEGID THN EF+ E
Sbjct 263 AKPFITSSKDFD-HLELRIAPELWLKRLIISGLQKVYEIGKVFRNEGIDSTHNAEFSTLE 321
Query 62 FYWAYADYNDTMRLTEDMISEMVMSIHGSYK 92
FY Y +D + TED+ ++ ++ ++
Sbjct 322 FYETYMSMDDIVTRTEDLFKFLITNLQKFFQ 352
> pfa:PF14_0166 lysine-tRNA ligase, putative (EC:6.1.1.6); K04567
lysyl-tRNA synthetase, class II [EC:6.1.1.6]
Length=674
Score = 82.4 bits (202), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 39/83 (46%), Positives = 54/83 (65%), Gaps = 1/83 (1%)
Query 2 AKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGM-DKVFEIGKNFRNEGIDMTHNPEFTAC 60
AKPF T+ LN LY+R++PEL+LK L V G+ +++FE+ K FRNEG+ HNPEFT
Sbjct 331 AKPFETYLKSLNLILYLRISPELFLKKLIVSGISEQIFELSKCFRNEGLSSIHNPEFTML 390
Query 61 EFYWAYADYNDTMRLTEDMISEM 83
E Y +Y +Y M E +I +
Sbjct 391 EIYKSYTNYKYMMNFVEKIIKHL 413
> tpv:TP04_0470 lysyl-tRNA synthetase; K04567 lysyl-tRNA synthetase,
class II [EC:6.1.1.6]
Length=708
Score = 81.3 bits (199), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 39/84 (46%), Positives = 57/84 (67%), Gaps = 1/84 (1%)
Query 2 AKPFVTHHNDLNTDLYMRVAPELYLKMLTVG-GMDKVFEIGKNFRNEGIDMTHNPEFTAC 60
A+PF T++ LN + +R+APE +K + +G + KVFEIGK FRNEG + H+PEFT
Sbjct 409 AEPFETYYKRLNEKVKLRIAPEFSIKKIFMGLPLTKVFEIGKCFRNEGSSLRHSPEFTMI 468
Query 61 EFYWAYADYNDTMRLTEDMISEMV 84
E Y ADYN ++L ED+I+ ++
Sbjct 469 EIYQKMADYNTMIKLLEDLINSLL 492
> tgo:TGME49_020350 lysyl-tRNA synthetase, putative (EC:6.1.1.12
6.1.1.6)
Length=1170
Score = 79.7 bits (195), Expect = 3e-15, Method: Composition-based stats.
Identities = 39/80 (48%), Positives = 54/80 (67%), Gaps = 1/80 (1%)
Query 2 AKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGM-DKVFEIGKNFRNEGIDMTHNPEFTAC 60
A+ F +H + L+ +++R+APEL LK L V G+ DKVFEIG+ FRNEGI H PEF +
Sbjct 743 ARLFESHLHALDLPVHLRIAPELALKRLVVSGVSDKVFEIGRCFRNEGISWRHQPEFLSM 802
Query 61 EFYWAYADYNDTMRLTEDMI 80
E Y + Y D M LTE+++
Sbjct 803 EAYATFWTYEDMMTLTEEIV 822
> ath:AT4G33760 tRNA synthetase class II (D, K and N) family protein
(EC:6.1.1.12); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=664
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 40/69 (57%), Gaps = 0/69 (0%)
Query 21 APELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACEFYWAYADYNDTMRLTEDMI 80
+P+L+ +ML V G DK ++I + FR+E + PEFT + A+ D ++L ED+I
Sbjct 275 SPQLFKQMLMVSGFDKYYQIARCFRDEDLRADRQPEFTQLDMEMAFMPMEDMLKLNEDLI 334
Query 81 SEMVMSIHG 89
++ I G
Sbjct 335 RKVFSEIKG 343
> eco:b4155 yjeA, ECK4151, genX, JW4116, poxA; EF-P-lysine34-lysine
ligase; K04568 lysyl-tRNA synthetase, class II [EC:6.1.1.6]
Length=325
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 46/80 (57%), Gaps = 0/80 (0%)
Query 5 FVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACEFYW 64
FV + +L++ +PE ++K L V G VF++ ++FRNE + HNPEFT E+Y
Sbjct 60 FVGPGHSQGMNLWLMTSPEYHMKRLLVAGCGPVFQLCRSFRNEEMGRYHNPEFTMLEWYR 119
Query 65 AYADYNDTMRLTEDMISEMV 84
+ D M +D++ +++
Sbjct 120 PHYDMYRLMNEVDDLLQQVL 139
> cel:F10C2.6 drs-2; aspartyl(D) tRNA Synthetase family member
(drs-2); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=593
Score = 50.1 bits (118), Expect = 3e-06, Method: Composition-based stats.
Identities = 21/60 (35%), Positives = 36/60 (60%), Gaps = 0/60 (0%)
Query 21 APELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACEFYWAYADYNDTMRLTEDMI 80
+P+ + ++L VG +D+ F+I + +R+EG PEFT + ++ N M+L EDMI
Sbjct 213 SPQQFKQLLMVGAIDRYFQIARCYRDEGSKGDRQPEFTQVDVEMSFTTQNGVMQLIEDMI 272
> cel:Y32F6A.6 hypothetical protein
Length=328
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 31/105 (29%), Positives = 47/105 (44%), Gaps = 16/105 (15%)
Query 12 LNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACEFYWAYADYND 71
+N D ELY +ML G+++VF+ P E + ND
Sbjct 131 INIDYVYSKMTELYNRMLVTQGIERVFKWNSQ---------RQPLVLTAEISMCNINLND 181
Query 72 TMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRL 116
L E++I+ +V SI S+ I + VE+DFTPP++RL
Sbjct 182 FQDLVEELITTLVYSIKRSFLIKYRS-------VEIDFTPPFKRL 219
> dre:571144 dars2, si:dkey-21n10.1; aspartyl-tRNA synthetase
2, mitochondrial; K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=660
Score = 48.9 bits (115), Expect = 7e-06, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 44/94 (46%), Gaps = 0/94 (0%)
Query 2 AKPFVTHHNDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACE 61
AK FV D + +P+ + ++L V G+D+ F++ + +R+EG PEFT +
Sbjct 244 AKEFVVPSGDPGKFYSLPQSPQQFKQLLMVAGIDRYFQLARCYRDEGSKPDRQPEFTQVD 303
Query 62 FYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPF 95
++ D M L E ++ G +PF
Sbjct 304 IEMSFVDQAGVMSLIEGLVQFSWPEDKGQINVPF 337
> sce:YPL104W MSD1, LPG5; Msd1p (EC:6.1.1.12); K01876 aspartyl-tRNA
synthetase [EC:6.1.1.12]
Length=658
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 42/79 (53%), Gaps = 1/79 (1%)
Query 4 PFVTHHNDLNTDLY-MRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACEF 62
P T +D Y + +P+ Y ++L G++K +++ + FR+E + PEFT +
Sbjct 206 PTRTKRSDGKPSFYALDQSPQQYKQLLMASGVNKYYQMARCFRDEDLRADRQPEFTQVDM 265
Query 63 YWAYADYNDTMRLTEDMIS 81
A+A+ D M++ E +S
Sbjct 266 EMAFANSEDVMKIIEKTVS 284
> mmu:226539 Dars2, 5830468K18Rik, MGC99963; aspartyl-tRNA synthetase
2 (mitochondrial) (EC:6.1.1.12); K01876 aspartyl-tRNA
synthetase [EC:6.1.1.12]
Length=653
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 35/60 (58%), Gaps = 0/60 (0%)
Query 21 APELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACEFYWAYADYNDTMRLTEDMI 80
+P+ + ++L VGG+D+ F++ + +R+EG PEFT + ++ + RL E ++
Sbjct 241 SPQQFKQLLMVGGLDRYFQVARCYRDEGSRPDRQPEFTQIDIEMSFVEQTGIQRLVEGLL 300
> hsa:55157 DARS2, ASPRS, FLJ10514, LBSL, MT-ASPRS, RP3-383J4.2;
aspartyl-tRNA synthetase 2, mitochondrial (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=645
Score = 43.1 bits (100), Expect = 3e-04, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 34/60 (56%), Gaps = 0/60 (0%)
Query 21 APELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACEFYWAYADYNDTMRLTEDMI 80
+P+ + ++L VGG+D+ F++ + +R+EG PEFT + ++ D L E ++
Sbjct 242 SPQQFKQLLMVGGLDRYFQVARCYRDEGSRPDRQPEFTQIDIEMSFVDQTGIQSLIEGLL 301
> tgo:TGME49_002530 aspartyl-tRNA synthetase, putative (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=773
Score = 42.0 bits (97), Expect = 8e-04, Method: Composition-based stats.
Identities = 33/125 (26%), Positives = 56/125 (44%), Gaps = 8/125 (6%)
Query 12 LNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDM-THNPEFTACEFYWAYAD-Y 69
N D + +P+LY +M ++VFEIG FR E + H EFT + + + Y
Sbjct 508 FNRDACLAQSPQLYKQMAMCADFERVFEIGPVFRAENSNTHRHLCEFTGLDLEMTFKEHY 567
Query 70 NDTMRLTEDMISEMVMSIHGSYKIP---FHPEGPNGPVVELDFTPPYRRLSLVEEIEKQA 126
++ + L +D+ + + K FH + P P ++ TP RLS E ++
Sbjct 568 SEVLDLLDDLFKFIFKGLSERCKKEIDLFHQQHPAEPFTWIEETP---RLSFEEGVQMLR 624
Query 127 KVTLP 131
+ P
Sbjct 625 EAGCP 629
> cel:B0464.1 drs-1; aspartyl(D) tRNA Synthetase family member
(drs-1); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=531
Score = 41.6 bits (96), Expect = 0.001, Method: Composition-based stats.
Identities = 25/98 (25%), Positives = 44/98 (44%), Gaps = 2/98 (2%)
Query 17 YMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDM-THNPEFTACEFYWAYA-DYNDTMR 74
Y+ +P+LY +M G +KV+ IG FR E + H EF + A+ Y++ M
Sbjct 275 YLAQSPQLYKQMAIAGDFEKVYTIGPVFRAEDSNTHRHMTEFVGLDLEMAFNFHYHEVME 334
Query 75 LTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPP 112
+++++M + +Y+ G P F P
Sbjct 335 TIAEVLTQMFKGLQQNYQDEIAAVGNQYPAEPFQFCEP 372
> pfa:PFA_0145c aspartyl-tRNA synthetase, putative (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=626
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 32/128 (25%), Positives = 57/128 (44%), Gaps = 7/128 (5%)
Query 10 NDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDM-THNPEFTACEFYWAYA- 67
N N + Y+ +P+LY +M G DKVFE+G FR E + H E+ + + Y
Sbjct 356 NYFNQNGYLAQSPQLYKQMCINSGFDKVFEVGPVFRAENSNTYRHLCEYVSLDIEMTYKF 415
Query 68 DYNDTMRLTEDMISEMVMSI-----HGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVEEI 122
DY + + + M + + + ++ + P+ V LD TP + ++ +
Sbjct 416 DYMENVHFYDSMFKHIFKELTNNEKNKTFIKTIKNQYPSDDFVWLDKTPIFTYEEAIKIL 475
Query 123 EKQAKVTL 130
K K+ L
Sbjct 476 IKNGKLFL 483
> bbo:BBOV_IV001690 21.m02784; aspartyl-tRNA synthetase (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=557
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 34/115 (29%), Positives = 50/115 (43%), Gaps = 6/115 (5%)
Query 12 LNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDM-THNPEFTACEFYWAYAD-Y 69
+ D + +P+LY ++ VG +VFEIG FR E + H EF + D Y
Sbjct 320 FDNDACLAQSPQLYKQLAVVGDFRRVFEIGPVFRAENSNTHRHLCEFVGLDMEMEIRDNY 379
Query 70 NDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVE----LDFTPPYRRLSLVE 120
+ + L +DM+ + I+ K + P VE LD TP R VE
Sbjct 380 MEVVDLIDDMLKHVFHGINTECKTEVEIFQKHQPSVEPFKWLDQTPRIRFTEAVE 434
> eco:b1866 aspS, ECK1867, JW1855, tls; aspartyl-tRNA synthetase
(EC:6.1.1.12); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=590
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 16/69 (23%), Positives = 36/69 (52%), Gaps = 0/69 (0%)
Query 21 APELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACEFYWAYADYNDTMRLTEDMI 80
+P+L+ ++L + G D+ ++I K FR+E + PEFT + ++ + E ++
Sbjct 193 SPQLFKQLLMMSGFDRYYQIVKCFRDEDLRADRQPEFTQIDVETSFMTAPQVREVMEALV 252
Query 81 SEMVMSIHG 89
+ + + G
Sbjct 253 RHLWLEVKG 261
> ath:AT4G31180 aspartyl-tRNA synthetase, putative / aspartate--tRNA
ligase, putative (EC:6.1.1.12); K01876 aspartyl-tRNA
synthetase [EC:6.1.1.12]
Length=558
Score = 38.5 bits (88), Expect = 0.009, Method: Composition-based stats.
Identities = 32/116 (27%), Positives = 52/116 (44%), Gaps = 12/116 (10%)
Query 21 APELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACEFYWAYAD------YNDTMR 74
+P+L+ +M G + +VFE+G FR E TH CEF + Y++ M
Sbjct 306 SPQLHKQMAICGDLRRVFEVGPVFRAED-SFTHR---HLCEFVGLDVEMEIRKHYSEIMD 361
Query 75 LTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVEEIE--KQAKV 128
L +++ + S++ K G P L F P RL+ E ++ K+A V
Sbjct 362 LVDELFVFIFTSLNERCKKELQAVGKQYPFEPLKFLPKTLRLTFEEGVQMLKEAGV 417
> hsa:1615 DARS, DKFZp781B11202, MGC111579; aspartyl-tRNA synthetase
(EC:6.1.1.12); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=501
Score = 37.7 bits (86), Expect = 0.013, Method: Composition-based stats.
Identities = 26/108 (24%), Positives = 43/108 (39%), Gaps = 2/108 (1%)
Query 17 YMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDM-THNPEFTACEFYWAYA-DYNDTMR 74
Y+ +P+LY +M +KVF IG FR E + H EF + A+ Y++ M
Sbjct 245 YLAQSPQLYKQMCICADFEKVFSIGPVFRAEDSNTHRHLTEFVGLDIEMAFNYHYHEVME 304
Query 75 LTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVEEI 122
D + ++ + ++ P F P RL E +
Sbjct 305 EIADTMVQIFKGLQERFQTEIQTVNKQFPCEPFKFLEPTLRLEYCEAL 352
> dre:324025 dars, MGC154056, wu:fc17a11, zgc:154056; aspartyl-tRNA
synthetase (EC:6.1.1.12); K01876 aspartyl-tRNA synthetase
[EC:6.1.1.12]
Length=531
Score = 37.0 bits (84), Expect = 0.023, Method: Composition-based stats.
Identities = 26/111 (23%), Positives = 44/111 (39%), Gaps = 2/111 (1%)
Query 12 LNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDM-THNPEFTACEFYWAYA-DY 69
T Y+ +P+LY +M DKVF +G FR E + H EF + A++ Y
Sbjct 270 FKTSAYLAQSPQLYKQMCICADFDKVFCVGPVFRAEDSNTHRHLTEFVGLDIEMAFSYHY 329
Query 70 NDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVE 120
++ + D + ++ + ++ P F P RL E
Sbjct 330 HEVIDSITDTMVQIFKGLRDRFQTEIQTVNKQYPSEPFKFLEPTLRLEYKE 380
> xla:380028 dars, MGC53970; aspartyl-tRNA synthetase (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=530
Score = 34.7 bits (78), Expect = 0.11, Method: Composition-based stats.
Identities = 26/113 (23%), Positives = 44/113 (38%), Gaps = 2/113 (1%)
Query 12 LNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDM-THNPEFTACEFYWAYA-DY 69
T Y+ +P+LY +M +KVF IG FR E + H EF + A+ Y
Sbjct 269 FKTSAYLAQSPQLYKQMCICADFEKVFCIGPVFRAEDSNTHRHLTEFVGLDIEMAFNYHY 328
Query 70 NDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVEEI 122
++ + D + ++ + ++ P F P RL E +
Sbjct 329 HEVVDEIADTLVQIFKGLQERFQTEIQTVCKQFPCEPFKFLEPTLRLEYTEGV 381
> xla:443916 dars, MGC80207; aspartyl-tRNA synthetase (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=530
Score = 34.7 bits (78), Expect = 0.12, Method: Composition-based stats.
Identities = 26/113 (23%), Positives = 44/113 (38%), Gaps = 2/113 (1%)
Query 12 LNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDM-THNPEFTACEFYWAYA-DY 69
T Y+ +P+LY +M +KVF IG FR E + H EF + A+ Y
Sbjct 269 FKTSAYLAQSPQLYKQMCICADFEKVFCIGPVFRAEDSNTHRHLTEFVGLDIEMAFNYHY 328
Query 70 NDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTPPYRRLSLVEEI 122
++ + D + ++ + ++ P F P RL E +
Sbjct 329 HEVVDEIADTMVQIFKGLQERFQTEIQTVCKQFPCEPFKFLEPTLRLEYTEGV 381
> ath:AT4G26870 aspartyl-tRNA synthetase, putative / aspartate--tRNA
ligase, putative (EC:6.1.1.12); K01876 aspartyl-tRNA
synthetase [EC:6.1.1.12]
Length=532
Score = 34.3 bits (77), Expect = 0.14, Method: Composition-based stats.
Identities = 32/109 (29%), Positives = 46/109 (42%), Gaps = 10/109 (9%)
Query 21 APELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACEFYWAYADYNDTMRLTE--D 78
+P+L+ +M G M +VFE+G FR E TH CEF + M +E D
Sbjct 280 SPQLHKQMAICGDMRRVFEVGPVFRAED-SFTHR---HLCEFVGLDVEMEIRMHYSEIMD 335
Query 79 MISEMVMSIHGSY--KIPFHPEG--PNGPVVELDFTPPYRRLSLVEEIE 123
++ E+ I + P E P L F P RL+ E I+
Sbjct 336 LVGELFPFIFTKIEERCPKELESVRKQYPFQSLKFLPQTLRLTFAEGIQ 384
> sce:YFL022C FRS2; Frs2p (EC:6.1.1.20); K01889 phenylalanyl-tRNA
synthetase alpha chain [EC:6.1.1.20]
Length=503
Score = 33.5 bits (75), Expect = 0.25, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 26/52 (50%), Gaps = 6/52 (11%)
Query 36 KVFEIGKNFRNEGIDMTHNPEFTACEFYWAYADYN----DTMRLTEDMISEM 83
++F I + FRNE +D TH EF E ADYN D ++ E+ M
Sbjct 351 RLFSIDRVFRNEAVDATHLAEFHQVE--GVLADYNITLGDLIKFMEEFFERM 400
> tpv:TP03_0489 aspartyl-tRNA synthetase (EC:6.1.1.12); K01876
aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=604
Score = 33.1 bits (74), Expect = 0.37, Method: Composition-based stats.
Identities = 23/78 (29%), Positives = 36/78 (46%), Gaps = 6/78 (7%)
Query 12 LNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACEFYWAYADYND 71
+ D + +P+LY +M G + +VFEIG FR E N CE+ +
Sbjct 285 FDQDACLAQSPQLYKQMAICGDLKRVFEIGPVFRAE----NSNTHRHLCEYVGLDMEMEI 340
Query 72 TMRLTE--DMISEMVMSI 87
TE D+I +M+ S+
Sbjct 341 LNDYTEVIDLIDQMLKSV 358
> cpv:cgd8_350 asparaginyl-tRNA synthetase (NOB+tRNA synthase)
; K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=499
Score = 32.7 bits (73), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 33/69 (47%), Gaps = 2/69 (2%)
Query 17 YMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMT-HNPEFTACEFYWAYADYNDTMRL 75
Y+ V+ +L L+ M V+ G FR E T H EF E A+AD +D M+L
Sbjct 240 YLTVSGQLALENFACS-MSDVYTFGPTFRAENSHTTRHLAEFWMIEPEMAFADLSDNMKL 298
Query 76 TEDMISEMV 84
E ++ V
Sbjct 299 GEGLLKYTV 307
> dre:568332 nars, fb57a10, si:dkey-274m14.2, wu:fb57a10; asparaginyl-tRNA
synthetase; K01893 asparaginyl-tRNA synthetase
[EC:6.1.1.22]
Length=556
Score = 32.7 bits (73), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 28/118 (23%), Positives = 52/118 (44%), Gaps = 11/118 (9%)
Query 10 NDLNTDLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNE-GIDMTHNPEFTACEFYWAYAD 68
N + Y+ + +LYL+ + + F I +++R E H E+T E +
Sbjct 296 NYFGENAYLTQSSQLYLET-CIPALGDTFCIAQSYRAEQSRTRRHLSEYTHIEAECPFMS 354
Query 69 YNDTMRLTEDMISEMVMSIHGSYKIPFHPEGPNGPVVELDFTP---PYRRLSLVEEIE 123
Y D + ED++ ++V + S P P + DF P P++R++ E I+
Sbjct 355 YEDLLNRLEDLVCDVVDRVLKS------PAAPLLYDINPDFKPPKRPFKRMNYTEAID 406
> cpv:cgd6_3390 membrane associated protein with 2 transmembrane
domains at the N-terminus and a phosphatidylinositol 4-kinase
domain at the C-terminus
Length=2158
Score = 31.2 bits (69), Expect = 1.3, Method: Composition-based stats.
Identities = 13/40 (32%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 56 EFTACEFYWAYADYNDTMRLTEDMISEMVMSIHGSYKIPF 95
++ E+ W Y +TM L E +S ++ SI+ S K+ F
Sbjct 1340 QYIVLEYIWEYFINGNTMILIEKYVSALIFSIYSSKKVGF 1379
> cpv:cgd7_1540 aspartate--tRNA ligase
Length=532
Score = 30.8 bits (68), Expect = 1.7, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 4/42 (9%)
Query 21 APELYLKMLTVGGMDKVFEIGKNFRNEGIDMTHNPEFTACEF 62
+P+L+ +M G +++VFEIG FR E N CEF
Sbjct 276 SPQLHKQMSICGDLERVFEIGPVFRAE----NSNTHRHLCEF 313
> xla:398071 mcm6, MGC85070, mis5, mmcm6; minichromosome maintenance
complex component 6 (EC:3.6.4.12); K02542 minichromosome
maintenance protein 6
Length=821
Score = 30.8 bits (68), Expect = 1.7, Method: Composition-based stats.
Identities = 21/83 (25%), Positives = 41/83 (49%), Gaps = 10/83 (12%)
Query 61 EFYWAYADYNDTMRLTEDMISEMVMSIHGSYKI----PFHPEGPNGPVVELDFTPPYRRL 116
EFY A++D+ ++ E +++ + S ++ P HPE +G + +D
Sbjct 111 EFYIAFSDFPARQKIRELSSAKIGTLLRISGQVVRTHPVHPELVSGTFLCMDCQ------ 164
Query 117 SLVEEIEKQAKVTLPRPLDGPEC 139
S+V+++E+Q + T P P C
Sbjct 165 SIVKDVEQQFRYTQPTICKNPVC 187
> pfa:PFE0715w asparagine-tRNA ligase, putative (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=1128
Score = 30.8 bits (68), Expect = 1.8, Method: Composition-based stats.
Identities = 20/79 (25%), Positives = 41/79 (51%), Gaps = 2/79 (2%)
Query 15 DLYMRVAPELYLKMLTVGGMDKVFEIGKNFRNEGIDMT-HNPEFTACEFYWA-YADYNDT 72
++ + +P+L+ +ML DKV+EI +RNE + H EF + + Y +Y +
Sbjct 611 NIVLSQSPQLFKQMLINSDFDKVYEINYCYRNEKFHSSRHLNEFMSLDIEKVIYDNYYEI 670
Query 73 MRLTEDMISEMVMSIHGSY 91
+ +++ +M I+ +Y
Sbjct 671 IIYMYNILKDMNTYINNNY 689
> dre:564982 mcm6l, MGC112244, zgc:112244; MCM6 minichromosome
maintenance deficient 6, like; K02542 minichromosome maintenance
protein 6
Length=824
Score = 30.4 bits (67), Expect = 2.5, Method: Composition-based stats.
Identities = 24/88 (27%), Positives = 39/88 (44%), Gaps = 20/88 (22%)
Query 61 EFYWAYADYNDTMRLTED---------MISEMVMSIHGSYKIPFHPEGPNGPVVELDFTP 111
EFY A++++ ++ E IS V+ H P HPE +G + LD
Sbjct 103 EFYVAFSEFPSRQKIRELSTVRIGTLLRISGQVVRTH-----PVHPELVSGTFLCLDCQ- 156
Query 112 PYRRLSLVEEIEKQAKVTLPRPLDGPEC 139
S+++++E+Q K T P P C
Sbjct 157 -----SVIKDVEQQFKYTQPTICKNPVC 179
Lambda K H
0.320 0.139 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3264639800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40