bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5447_orf1
Length=187
Score E
Sequences producing significant alignments: (Bits) Value
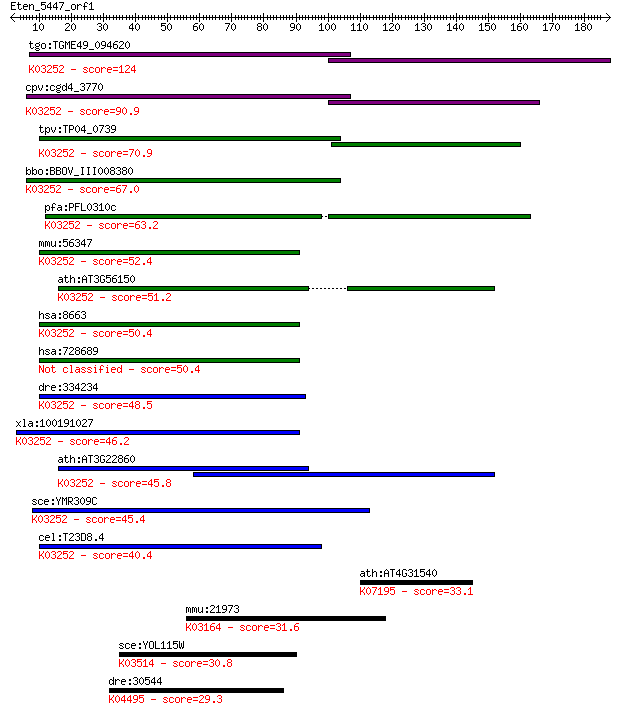
tgo:TGME49_094620 eukaryotic translation initiation factor 3 s... 124 2e-28
cpv:cgd4_3770 hypothetical protein ; K03252 translation initia... 90.9 2e-18
tpv:TP04_0739 eukaryotic translation initiation factor 3 subun... 70.9 2e-12
bbo:BBOV_III008380 17.m07733; eukaryotic translation initiatio... 67.0 3e-11
pfa:PFL0310c eukaryotic translation initiation factor 3 subuni... 63.2 5e-10
mmu:56347 Eif3c, 110kDa, 3230401O13Rik, Eif3s8, MGC36637, NIPI... 52.4 1e-06
ath:AT3G56150 EIF3C; EIF3C (EUKARYOTIC TRANSLATION INITIATION ... 51.2 2e-06
hsa:8663 EIF3C, EIF3CL, EIF3S8, FLJ53378, FLJ54400, FLJ54404, ... 50.4 4e-06
hsa:728689 EIF3CL; eukaryotic translation initiation factor 3,... 50.4 4e-06
dre:334234 eif3c, eif3s8, fi37c04, wu:fb34a08, wu:fi37c04, zgc... 48.5 1e-05
xla:100191027 eif3c, MGC114671, eif3-p110, eif3s8; eukaryotic ... 46.2 6e-05
ath:AT3G22860 TIF3C2; TIF3C2; translation initiation factor; K... 45.8 9e-05
sce:YMR309C NIP1; Nip1p; K03252 translation initiation factor ... 45.4 1e-04
cel:T23D8.4 eif-3.C; Eukaryotic Initiation Factor family membe... 40.4 0.004
ath:AT4G31540 ATEXO70G1 (exocyst subunit EXO70 family protein ... 33.1 0.55
mmu:21973 Top2a, Top-2; topoisomerase (DNA) II alpha (EC:5.99.... 31.6 1.7
sce:YOL115W PAP2, TRF4; Non-canonical poly(A) polymerase, invo... 30.8 3.2
dre:30544 sox17, MGC91776; SRY-box containing gene 17; K04495 ... 29.3 7.9
> tgo:TGME49_094620 eukaryotic translation initiation factor 3
subunit 8, putative ; K03252 translation initiation factor
3 subunit C
Length=1104
Score = 124 bits (310), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 59/100 (59%), Positives = 79/100 (79%), Gaps = 0/100 (0%)
Query 7 EIQQMSASILTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFLDERG 66
+ Q +S+ ILTSFVERLDD+L KSLQ TDVHS+EYKERLG+S+D++A+L R + L +
Sbjct 436 DAQVVSSGILTSFVERLDDELMKSLQFTDVHSEEYKERLGQSVDMIAVLCRAWVHLTDCQ 495
Query 67 FKAQAATVALKVNEHMHYKPDTIASKMWDVLRKELAETVS 106
K QAAT+AL++NEHMHYK D IA+ MWD++RK + V+
Sbjct 496 SKPQAATLALRINEHMHYKHDAIAASMWDLVRKRVPAEVA 535
Score = 67.4 bits (163), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 47/107 (43%), Positives = 59/107 (55%), Gaps = 20/107 (18%)
Query 100 ELAETVSPQCHLEVLGHLISAEFDTTSGVFTSMPLQIWMETFAGVKLLLSIMEQ------ 153
ELA V PQ LEVL HLISAEFDTTSGVF + IW++ F ++ +L +E+
Sbjct 345 ELAARVGPQSELEVLAHLISAEFDTTSGVFACLSSAIWLDVFHQLEAVLGHLEEGVKKAR 404
Query 154 --NKGA-----YLVPLAGLAESSTPE------EIQQMSASILTSFVE 187
KGA LVP LA S + + Q +S+ ILTSFVE
Sbjct 405 ADGKGAGGTAYVLVPTV-LASSDAVDAALVAPDAQVVSSGILTSFVE 450
> cpv:cgd4_3770 hypothetical protein ; K03252 translation initiation
factor 3 subunit C
Length=1040
Score = 90.9 bits (224), Expect = 2e-18, Method: Composition-based stats.
Identities = 41/101 (40%), Positives = 71/101 (70%), Gaps = 0/101 (0%)
Query 6 EEIQQMSASILTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFLDER 65
+E + + + L SF+ERLD + K+LQ TDVHS EYK+RL +S+ +LALLWR + +ER
Sbjct 472 KEQDKSTVTFLVSFIERLDGESLKALQLTDVHSSEYKDRLVQSLHLLALLWRCYKICEER 531
Query 66 GFKAQAATVALKVNEHMHYKPDTIASKMWDVLRKELAETVS 106
G+ +++++ + +H+K D++A K+W+ +R+ L +TV+
Sbjct 532 GYYDLVSSLSVHLINQLHFKNDSLAIKVWEFVRQILEKTVN 572
Score = 44.3 bits (103), Expect = 3e-04, Method: Composition-based stats.
Identities = 23/66 (34%), Positives = 37/66 (56%), Gaps = 0/66 (0%)
Query 100 ELAETVSPQCHLEVLGHLISAEFDTTSGVFTSMPLQIWMETFAGVKLLLSIMEQNKGAYL 159
E++ +S Q + +VL HLISA+FDT +G F + IW E + +LL ++ N
Sbjct 354 EISRPLSLQAYADVLTHLISAQFDTITGAFHCITSGIWSEICDNINILLDLVLLNNKDKR 413
Query 160 VPLAGL 165
V ++G
Sbjct 414 VYISGF 419
> tpv:TP04_0739 eukaryotic translation initiation factor 3 subunit
8; K03252 translation initiation factor 3 subunit C
Length=951
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/96 (38%), Positives = 65/96 (67%), Gaps = 3/96 (3%)
Query 10 QMSASILTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFL--DERGF 67
+ S ++L++ V++L+D+L K L T+VH+ +YK L +ID+L LL RT + E+G
Sbjct 459 KTSLTVLSTIVQKLNDELYKGLLYTEVHNPDYKTMLAYTIDMLYLLHRTLVYYLKFEKGN 518
Query 68 KAQAATVALKVNEHMHYKPDTIASKMWDVLRKELAE 103
+ AAT AL + +H HYK D I++K+W+++R ++ +
Sbjct 519 EF-AATTALMILDHCHYKDDEISTKIWELVRNKIKD 553
Score = 34.7 bits (78), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 31/59 (52%), Gaps = 0/59 (0%)
Query 101 LAETVSPQCHLEVLGHLISAEFDTTSGVFTSMPLQIWMETFAGVKLLLSIMEQNKGAYL 159
+A+T+S +LEVL L+ FD S + +M W+ T+ L+S + N +YL
Sbjct 358 IAKTISQSLYLEVLETLVHVLFDNYSHAYGAMTPTEWINTYRIASHLVSELISNPKSYL 416
> bbo:BBOV_III008380 17.m07733; eukaryotic translation initiation
factor 3 subunit 8; K03252 translation initiation factor
3 subunit C
Length=966
Score = 67.0 bits (162), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 63/100 (63%), Gaps = 2/100 (2%)
Query 6 EEIQQMSASILTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFLDER 65
EE Q S +IL S V +L+D+L K L +V SD+Y L ++++L LL +T + +
Sbjct 476 EERIQRSMTILESVVRKLNDELYKGLLYIEVGSDDYNTMLVYNVNMLYLLHKTLLYCLSK 535
Query 66 GFK--AQAATVALKVNEHMHYKPDTIASKMWDVLRKELAE 103
G + AA +A+ + EH+HYK D ++ K+W+++R+++ +
Sbjct 536 GKETITHAANIAIIMLEHLHYKDDVVSGKIWELVRQKVTD 575
> pfa:PFL0310c eukaryotic translation initiation factor 3 subunit
8, putative; K03252 translation initiation factor 3 subunit
C
Length=984
Score = 63.2 bits (152), Expect = 5e-10, Method: Composition-based stats.
Identities = 30/63 (47%), Positives = 43/63 (68%), Gaps = 0/63 (0%)
Query 100 ELAETVSPQCHLEVLGHLISAEFDTTSGVFTSMPLQIWMETFAGVKLLLSIMEQNKGAYL 159
E+A+T+S Q ++EVL HLI+ EFD S V+T M IW + F V+L+L I+ QN+ YL
Sbjct 382 EIAKTISTQSYIEVLEHLINLEFDVVSSVYTYMSFNIWNKVFKYVELILDILIQNENFYL 441
Query 160 VPL 162
V +
Sbjct 442 VSI 444
Score = 62.8 bits (151), Expect = 6e-10, Method: Composition-based stats.
Identities = 30/87 (34%), Positives = 55/87 (63%), Gaps = 1/87 (1%)
Query 12 SASILTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFLD-ERGFKAQ 70
S L SF+ +LDD+L K+L DV ++EY++RLGK+I ++ LL + + ++ +
Sbjct 464 SCKTLISFLAKLDDELLKALLYIDVQTEEYRKRLGKTIHMIGLLKKGYNYVKCLKNMPDL 523
Query 71 AATVALKVNEHMHYKPDTIASKMWDVL 97
A ++ ++ EHM+YKP+ + ++W L
Sbjct 524 AIHISSRILEHMYYKPEMLFKQIWTYL 550
> mmu:56347 Eif3c, 110kDa, 3230401O13Rik, Eif3s8, MGC36637, NIPIL(A3),
NipilA3; eukaryotic translation initiation factor 3,
subunit C; K03252 translation initiation factor 3 subunit
C
Length=911
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 46/81 (56%), Gaps = 1/81 (1%)
Query 10 QMSASILTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKA 69
++ ILT VER+D++ TK +Q+TD HS EY E L + A++ R +L+E+G
Sbjct 437 RVRGCILT-LVERMDEEFTKIMQNTDPHSQEYVEHLKDEAQVCAIIERVQRYLEEKGTTE 495
Query 70 QAATVALKVNEHMHYKPDTIA 90
+ + L+ H +YK D A
Sbjct 496 EICQIYLRRILHTYYKFDYKA 516
> ath:AT3G56150 EIF3C; EIF3C (EUKARYOTIC TRANSLATION INITIATION
FACTOR 3C); translation initiation factor; K03252 translation
initiation factor 3 subunit C
Length=900
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/79 (41%), Positives = 44/79 (55%), Gaps = 2/79 (2%)
Query 16 LTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFLDERG-FKAQAATV 74
L +F+ER+D + KSLQ D H+ EY ERL LAL + + G FKA AA V
Sbjct 356 LVAFLERVDTEFFKSLQCIDPHTREYVERLRDEPMFLALAQNIQDYFERMGDFKA-AAKV 414
Query 75 ALKVNEHMHYKPDTIASKM 93
AL+ E ++YKP + M
Sbjct 415 ALRRVEAIYYKPQEVYDAM 433
Score = 36.6 bits (83), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 106 SPQCHLEVLGHLISAEFDTTSGVFTSMPLQIWMETFAGVKLLLSIM 151
+P LE+L +ISA+FD G+ MP+ +W + + +L I+
Sbjct 276 TPAQKLEILFSVISAQFDVNPGLSGHMPINVWKKCVLNMLTILDIL 321
> hsa:8663 EIF3C, EIF3CL, EIF3S8, FLJ53378, FLJ54400, FLJ54404,
FLJ55450, FLJ55750, FLJ78287, MGC189737, MGC189744, eIF3-p110;
eukaryotic translation initiation factor 3, subunit C;
K03252 translation initiation factor 3 subunit C
Length=913
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 45/81 (55%), Gaps = 1/81 (1%)
Query 10 QMSASILTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKA 69
++ ILT VER+D++ TK +Q+TD HS EY E L + A++ R +L+E+G
Sbjct 439 RVRGCILT-LVERMDEEFTKIMQNTDPHSQEYVEHLKDEAQVCAIIERVQRYLEEKGTTE 497
Query 70 QAATVALKVNEHMHYKPDTIA 90
+ + L H +YK D A
Sbjct 498 EVCRIYLLRILHTYYKFDYKA 518
> hsa:728689 EIF3CL; eukaryotic translation initiation factor
3, subunit C-like
Length=914
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 45/81 (55%), Gaps = 1/81 (1%)
Query 10 QMSASILTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKA 69
++ ILT VER+D++ TK +Q+TD HS EY E L + A++ R +L+E+G
Sbjct 440 RVRGCILT-LVERMDEEFTKIMQNTDPHSQEYVEHLKDEAQVCAIIERVQRYLEEKGTTE 498
Query 70 QAATVALKVNEHMHYKPDTIA 90
+ + L H +YK D A
Sbjct 499 EVCRIYLLRILHTYYKFDYKA 519
> dre:334234 eif3c, eif3s8, fi37c04, wu:fb34a08, wu:fi37c04, zgc:66128;
eukaryotic translation initiation factor 3, subunit
C; K03252 translation initiation factor 3 subunit C
Length=926
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/83 (32%), Positives = 45/83 (54%), Gaps = 1/83 (1%)
Query 10 QMSASILTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKA 69
++ ILT VER+D++ TK +Q+TD HS EY + L + ++ R +L+ +G
Sbjct 439 RVRGCILT-LVERMDEEFTKIMQNTDPHSQEYVDNLKDEGRVCGIIDRLLQYLETKGSTE 497
Query 70 QAATVALKVNEHMHYKPDTIASK 92
+ V L+ H +YK D A +
Sbjct 498 EVCRVYLRRIMHTYYKFDYKAHR 520
> xla:100191027 eif3c, MGC114671, eif3-p110, eif3s8; eukaryotic
translation initiation factor 3, subunit C; K03252 translation
initiation factor 3 subunit C
Length=926
Score = 46.2 bits (108), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 47/88 (53%), Gaps = 1/88 (1%)
Query 3 STPEEIQQMSASILTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFL 62
S E+ ++ ILT +ER++++ TK +Q+TD HS EY + L + ++ R +L
Sbjct 435 SNLEQPLRVRGCILT-LIERMEEEFTKIMQNTDPHSQEYVDNLKDEARVCEVIERAQKYL 493
Query 63 DERGFKAQAATVALKVNEHMHYKPDTIA 90
E+G + V L+ H +YK D A
Sbjct 494 QEKGSTEEICRVYLRRIMHTYYKFDYKA 521
> ath:AT3G22860 TIF3C2; TIF3C2; translation initiation factor;
K03252 translation initiation factor 3 subunit C
Length=805
Score = 45.8 bits (107), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 42/78 (53%), Gaps = 0/78 (0%)
Query 16 LTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKAQAATVA 75
L +F+E+++ + KSLQ D H+++Y ERL LAL +L+ G A+ VA
Sbjct 318 LVAFLEKIETEFFKSLQCIDPHTNDYVERLKDEPMFLALAQSIQDYLERTGDSKAASKVA 377
Query 76 LKVNEHMHYKPDTIASKM 93
+ E ++YKP + M
Sbjct 378 FILVESIYYKPQEVFDAM 395
Score = 34.7 bits (78), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 53/96 (55%), Gaps = 6/96 (6%)
Query 58 TFCFLDERGFKAQAATVALKVNEHMHYKP-DTIASKMWDVLRKELAETVSPQCHLEVLGH 116
T+ ++++ + +AA + + + + KP +T A K D+ + +A+T P +E+L
Sbjct 192 TWNMVNKKFKEIRAARWSKRRSSSLKLKPGETHAQKHMDLTK--IAKT--PAQKVEILFS 247
Query 117 LISAEFDTTSGVFTS-MPLQIWMETFAGVKLLLSIM 151
+ISAEF+ SG + MP+ +W + + +L I+
Sbjct 248 VISAEFNVNSGGLSGYMPIDVWKKCVVNMLTILDIL 283
> sce:YMR309C NIP1; Nip1p; K03252 translation initiation factor
3 subunit C
Length=812
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 32/114 (28%), Positives = 55/114 (48%), Gaps = 10/114 (8%)
Query 8 IQQMSASILTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFL----- 62
++++ SI SFVERLDD+ KSL + D HS +Y RL I L+ RT +
Sbjct 362 VKRILGSIF-SFVERLDDEFMKSLLNIDPHSSDYLIRLRDEQSIYNLILRTQLYFEATLK 420
Query 63 DERGFKAQAATVALKVNEHMHYKPDTIASKM----WDVLRKELAETVSPQCHLE 112
DE + +K +H++YK + + M W+++ + + + L+
Sbjct 421 DEHDLERALTRPFVKRLDHIYYKSENLIKIMETAAWNIIPAQFKSKFTSKDQLD 474
> cel:T23D8.4 eif-3.C; Eukaryotic Initiation Factor family member
(eif-3.C); K03252 translation initiation factor 3 subunit
C
Length=898
Score = 40.4 bits (93), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 51/97 (52%), Gaps = 10/97 (10%)
Query 10 QMSASILTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFLDERG--- 66
++ SIL + V+RLD +L K LQ+ D HS++Y E+L D+ +L+ + +++ R
Sbjct 414 RIQGSILIA-VQRLDGELAKILQNADCHSNDYIEKLKAEKDMCSLIEKAEKYVELRNDSG 472
Query 67 --FKAQAATVALKVNEHMHYK----PDTIASKMWDVL 97
K + V + EH +YK + A K+ D L
Sbjct 473 IFDKHEVCKVYMMRIEHAYYKYQDQNEEDAGKLMDYL 509
> ath:AT4G31540 ATEXO70G1 (exocyst subunit EXO70 family protein
G1); protein binding; K07195 exocyst complex component 7
Length=687
Score = 33.1 bits (74), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 21/36 (58%), Gaps = 1/36 (2%)
Query 110 HLE-VLGHLISAEFDTTSGVFTSMPLQIWMETFAGV 144
HLE + HL AEF + VF + L +WM+ F+ +
Sbjct 299 HLEFAVKHLFEAEFKLCNDVFERLGLNVWMDCFSKI 334
> mmu:21973 Top2a, Top-2; topoisomerase (DNA) II alpha (EC:5.99.1.3);
K03164 DNA topoisomerase II [EC:5.99.1.3]
Length=1528
Score = 31.6 bits (70), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 33/65 (50%), Gaps = 4/65 (6%)
Query 56 WRTFCFLDERGFKAQAATVAL---KVNEHMHYKPDTIASKMWDVLRKELAETVSPQCHLE 112
W + ERGF+ + ++ K H+ Y D I SK+ DV++K+ V+ + H +
Sbjct 296 WEVCLTMSERGFQQISFVNSIATSKGGRHVDYVADQIVSKLVDVVKKKNKGGVAVKAH-Q 354
Query 113 VLGHL 117
V H+
Sbjct 355 VKNHM 359
> sce:YOL115W PAP2, TRF4; Non-canonical poly(A) polymerase, involved
in nuclear RNA degradation as a component of the TRAMP
complex; catalyzes polyadenylation of hypomodified tRNAs,
and snoRNA and rRNA precursors; overlapping but non-redundant
functions with Trf5p (EC:2.7.7.-); K03514 DNA polymerase sigma
subunit [EC:2.7.7.7]
Length=584
Score = 30.8 bits (68), Expect = 3.2, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 30/57 (52%), Gaps = 5/57 (8%)
Query 35 DVHSDEYKERLGKSI--DILALLWRTFCFLDERGFKAQAATVALKVNEHMHYKPDTI 89
++HS +K+RLGKSI +++ + F DERG A + NE+ H K I
Sbjct 460 ELHSATFKDRLGKSILGNVIKYRGKARDFKDERGLVLNKAIIE---NENYHKKRSRI 513
> dre:30544 sox17, MGC91776; SRY-box containing gene 17; K04495
transcription factor SOX17 (SOX group F)
Length=413
Score = 29.3 bits (64), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 30/55 (54%), Gaps = 8/55 (14%)
Query 32 QSTDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKAQAATVALK-VNEHMHYK 85
Q+ D+H+ E + LGKS W+ +D+R F +A + +K + +H +YK
Sbjct 84 QNPDLHNAELSKMLGKS-------WKALPMVDKRPFVEEAERLRVKHMQDHPNYK 131
Lambda K H
0.316 0.129 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5235419772
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40