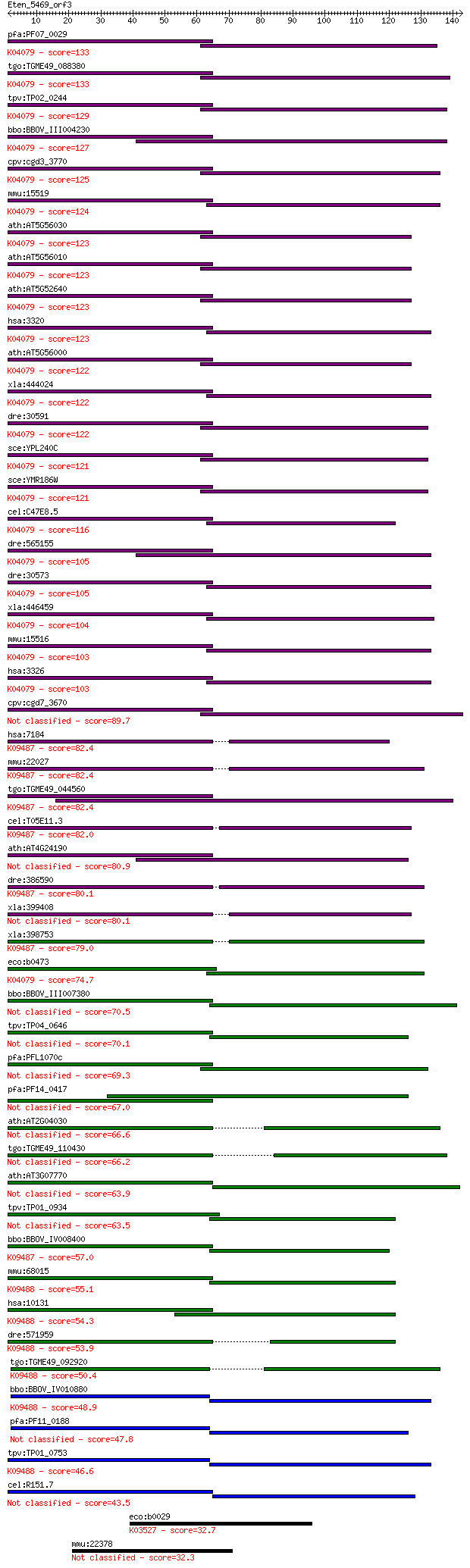
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5469_orf3
Length=142
Score E
Sequences producing significant alignments: (Bits) Value
pfa:PF07_0029 heat shock protein 86; K04079 molecular chaperon... 133 1e-31
tgo:TGME49_088380 heat shock protein 90 ; K04079 molecular cha... 133 1e-31
tpv:TP02_0244 heat shock protein 90; K04079 molecular chaperon... 129 3e-30
bbo:BBOV_III004230 17.m07381; hsp90 protein; K04079 molecular ... 127 2e-29
cpv:cgd3_3770 Hsp90 ; K04079 molecular chaperone HtpG 125 4e-29
mmu:15519 Hsp90aa1, 86kDa, 89kDa, AL024080, AL024147, Hsp86-1,... 124 1e-28
ath:AT5G56030 HSP81-2 (HEAT SHOCK PROTEIN 81-2); ATP binding; ... 123 1e-28
ath:AT5G56010 HSP81-3; ATP binding / unfolded protein binding;... 123 2e-28
ath:AT5G52640 ATHSP90.1 (HEAT SHOCK PROTEIN 90.1); ATP binding... 123 2e-28
hsa:3320 HSP90AA1, FLJ31884, HSP86, HSP89A, HSP90A, HSP90N, HS... 123 2e-28
ath:AT5G56000 heat shock protein 81-4 (HSP81-4); K04079 molecu... 122 3e-28
xla:444024 hsp90aa1.1, MGC82579, hsp86, hsp89, hsp90, hsp90a, ... 122 5e-28
dre:30591 hsp90a.1, fb17b01, hsp90, hsp90a, hsp90alpha, wu:fb1... 122 5e-28
sce:YPL240C HSP82, HSP90; Hsp82p; K04079 molecular chaperone HtpG 121 8e-28
sce:YMR186W HSC82, HSP90; Cytoplasmic chaperone of the Hsp90 f... 121 9e-28
cel:C47E8.5 daf-21; abnormal DAuer Formation family member (da... 116 2e-26
dre:565155 hsp90a.2, cb820, hsp90a2; heat shock protein 90-alp... 105 3e-23
dre:30573 hsp90ab1, hsp90b, hsp90beta, wu:fa29f01, wu:fa91e11,... 105 4e-23
xla:446459 hsp90ab1, hsp90b, hsp90beta; heat shock protein 90k... 104 1e-22
mmu:15516 Hsp90ab1, 90kDa, AL022974, C81438, Hsp84, Hsp84-1, H... 103 1e-22
hsa:3326 HSP90AB1, D6S182, FLJ26984, HSP90-BETA, HSP90B, HSPC2... 103 1e-22
cpv:cgd7_3670 heat shock protein 90 (Hsp90), signal peptide pl... 89.7 3e-18
hsa:7184 HSP90B1, ECGP, GP96, GRP94, TRA1; heat shock protein ... 82.4 4e-16
mmu:22027 Hsp90b1, ERp99, GRP94, TA-3, Targ2, Tra-1, Tra1, end... 82.4 4e-16
tgo:TGME49_044560 heat shock protein 90, putative (EC:2.7.13.3... 82.4 5e-16
cel:T05E11.3 hypothetical protein; K09487 heat shock protein 9... 82.0 5e-16
ath:AT4G24190 SHD; SHD (SHEPHERD); ATP binding / unfolded prot... 80.9 1e-15
dre:386590 hsp90b1, GP96, GRP94, fb61d09, tra-1, tra1, wu:fb61... 80.1 2e-15
xla:399408 hsp90b1, ecgp, gp96, grp94, tra1; heat shock protei... 80.1 2e-15
xla:398753 hypothetical protein MGC68448; K09487 heat shock pr... 79.0 4e-15
eco:b0473 htpG, ECK0467, JW0462; molecular chaperone HSP90 fam... 74.7 9e-14
bbo:BBOV_III007380 17.m07646; heat shock protein 90 70.5
tpv:TP04_0646 heat shock protein 90 70.1
pfa:PFL1070c endoplasmin homolog precursor, putative 69.3 3e-12
pfa:PF14_0417 HSP90 67.0 2e-11
ath:AT2G04030 CR88; CR88; ATP binding 66.6 2e-11
tgo:TGME49_110430 heat shock protein 90, putative (EC:3.2.1.3 ... 66.2 3e-11
ath:AT3G07770 ATP binding 63.9 1e-10
tpv:TP01_0934 heat shock protein 90 63.5
bbo:BBOV_IV008400 23.m06066; heat shock protein 90; K09487 hea... 57.0 2e-08
mmu:68015 Trap1, 2410002K23Rik, HSP75; TNF receptor-associated... 55.1 8e-08
hsa:10131 TRAP1, HSP75, HSP90L; TNF receptor-associated protei... 54.3 1e-07
dre:571959 trap1, fc85a11, wu:fc85a11; TNF receptor-associated... 53.9 2e-07
tgo:TGME49_092920 heat shock protein 90, putative ; K09488 TNF... 50.4 2e-06
bbo:BBOV_IV010880 23.m05763; heat shock protein 75; K09488 TNF... 48.9 6e-06
pfa:PF11_0188 heat shock protein 90, putative 47.8 1e-05
tpv:TP01_0753 heat shock protein 75; K09488 TNF receptor-assoc... 46.6 3e-05
cel:R151.7 hypothetical protein 43.5 2e-04
eco:b0029 ispH, ECK0030, JW0027, lytB, yaaE; 4-hydroxy-3-methy... 32.7 0.38
mmu:22378 Wbp2; WW domain binding protein 2 32.3
> pfa:PF07_0029 heat shock protein 86; K04079 molecular chaperone
HtpG
Length=745
Score = 133 bits (335), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 59/64 (92%), Positives = 62/64 (96%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEFKALLF+PKRAPFD+FE RKKRNNIKLYVRR FIMDDCE+IIPEWLNFVKG
Sbjct 347 HFSVEGQLEFKALLFIPKRAPFDMFENRKKRNNIKLYVRRVFIMDDCEEIIPEWLNFVKG 406
Query 61 VVDS 64
VVDS
Sbjct 407 VVDS 410
Score = 110 bits (276), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 54/74 (72%), Positives = 64/74 (86%), Gaps = 0/74 (0%)
Query 61 VVDSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIE 120
V +SAAGGSFTV KD+ E LGRGT+IILHLKEDQ EYLEE+R+KDLVKKHSEFISFPI+
Sbjct 147 VWESAAGGSFTVTKDETNEKLGRGTKIILHLKEDQLEYLEEKRIKDLVKKHSEFISFPIK 206
Query 121 LAVEKTHEREVTES 134
L E+ +E+E+T S
Sbjct 207 LYCERQNEKEITAS 220
> tgo:TGME49_088380 heat shock protein 90 ; K04079 molecular chaperone
HtpG
Length=708
Score = 133 bits (335), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 58/64 (90%), Positives = 63/64 (98%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEFKALLF+PKRAPFDLFETRKKRNN++LYVRR FIMDDCED+IPEWLNFV+G
Sbjct 305 HFSVEGQLEFKALLFLPKRAPFDLFETRKKRNNVRLYVRRVFIMDDCEDLIPEWLNFVRG 364
Query 61 VVDS 64
VVDS
Sbjct 365 VVDS 368
Score = 116 bits (290), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 59/79 (74%), Positives = 71/79 (89%), Gaps = 1/79 (1%)
Query 61 VVDSAAGGSFTVQK-DDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPI 119
V +S+AGGSFTV K + ++E + RGTRIILH+KEDQ EYLE+RRLKDLVKKHSEFISFPI
Sbjct 148 VWESSAGGSFTVSKAEGQFENIVRGTRIILHMKEDQTEYLEDRRLKDLVKKHSEFISFPI 207
Query 120 ELAVEKTHEREVTESEDEE 138
ELAVEK+ ++E+TESEDEE
Sbjct 208 ELAVEKSVDKEITESEDEE 226
> tpv:TP02_0244 heat shock protein 90; K04079 molecular chaperone
HtpG
Length=721
Score = 129 bits (324), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 56/64 (87%), Positives = 63/64 (98%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEFKALLFVP+RAPFD+FE+RKK+NNIKLYVRR FIMDDCE++IPEWL+FVKG
Sbjct 323 HFSVEGQLEFKALLFVPRRAPFDMFESRKKKNNIKLYVRRVFIMDDCEELIPEWLSFVKG 382
Query 61 VVDS 64
VVDS
Sbjct 383 VVDS 386
Score = 117 bits (292), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 58/77 (75%), Positives = 65/77 (84%), Gaps = 0/77 (0%)
Query 61 VVDSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIE 120
V +S A G FTV+KDD +EPL RGTR+ILHLKEDQ EYLEERRLK+LVKKHSEFISFPI
Sbjct 154 VWESTASGHFTVKKDDSHEPLKRGTRLILHLKEDQTEYLEERRLKELVKKHSEFISFPIS 213
Query 121 LAVEKTHEREVTESEDE 137
L+VEKT E EVT+ E E
Sbjct 214 LSVEKTQETEVTDDEAE 230
> bbo:BBOV_III004230 17.m07381; hsp90 protein; K04079 molecular
chaperone HtpG
Length=712
Score = 127 bits (318), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 55/64 (85%), Positives = 61/64 (95%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEFKA+LFVPKRAPFD+FE RKK+NNIKLYVRR FIMDDC+++IPEWL FVKG
Sbjct 315 HFSVEGQLEFKAILFVPKRAPFDMFENRKKKNNIKLYVRRVFIMDDCDELIPEWLGFVKG 374
Query 61 VVDS 64
VVDS
Sbjct 375 VVDS 378
Score = 106 bits (265), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 57/97 (58%), Positives = 72/97 (74%), Gaps = 0/97 (0%)
Query 41 AFIMDDCEDIIPEWLNFVKGVVDSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLE 100
A+++ D ++ + N + V +S A G FTV KD+ + L RGTR+ILHLK+DQ EYLE
Sbjct 130 AYLVADKVTVVSKNNNDDQYVWESNASGHFTVTKDESEDQLKRGTRLILHLKDDQSEYLE 189
Query 101 ERRLKDLVKKHSEFISFPIELAVEKTHEREVTESEDE 137
ERRLK+LVKKHSEFISFPI L+VEKT E EVT+ E E
Sbjct 190 ERRLKELVKKHSEFISFPIRLSVEKTTETEVTDDEAE 226
> cpv:cgd3_3770 Hsp90 ; K04079 molecular chaperone HtpG
Length=711
Score = 125 bits (314), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 55/64 (85%), Positives = 62/64 (96%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEFKA+LF+P+RAPFDLFETRKKRNNIKLYVRR FIMDDCE++IPE+L FV+G
Sbjct 312 HFSVEGQLEFKAILFIPRRAPFDLFETRKKRNNIKLYVRRVFIMDDCEELIPEFLGFVRG 371
Query 61 VVDS 64
VVDS
Sbjct 372 VVDS 375
Score = 108 bits (269), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 52/75 (69%), Positives = 63/75 (84%), Gaps = 0/75 (0%)
Query 61 VVDSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIE 120
+ +S+AGGSFT+ D L RGTRIILHLKEDQ +YLEER L+DLVKKHSEFISFPIE
Sbjct 158 IWESSAGGSFTITNDTSDNKLQRGTRIILHLKEDQLDYLEERTLRDLVKKHSEFISFPIE 217
Query 121 LAVEKTHEREVTESE 135
L+VEKT E+E+T+S+
Sbjct 218 LSVEKTTEKEITDSD 232
> mmu:15519 Hsp90aa1, 86kDa, 89kDa, AL024080, AL024147, Hsp86-1,
Hsp89, Hsp90, Hspca, hsp4; heat shock protein 90, alpha (cytosolic),
class A member 1; K04079 molecular chaperone HtpG
Length=733
Score = 124 bits (310), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 53/64 (82%), Positives = 62/64 (96%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEF+ALLFVP+RAPFDLFE RKK+NNIKLYVRR FIMD+CE++IPE+LNF++G
Sbjct 329 HFSVEGQLEFRALLFVPRRAPFDLFENRKKKNNIKLYVRRVFIMDNCEELIPEYLNFIRG 388
Query 61 VVDS 64
VVDS
Sbjct 389 VVDS 392
Score = 99.4 bits (246), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 47/73 (64%), Positives = 62/73 (84%), Gaps = 1/73 (1%)
Query 63 DSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELA 122
+S+AGGSFTV+ D EP+GRGT++ILHLKEDQ EYLEERR+K++VKKHS+FI +PI L
Sbjct 163 ESSAGGSFTVRTDTG-EPMGRGTKVILHLKEDQTEYLEERRIKEIVKKHSQFIGYPITLF 221
Query 123 VEKTHEREVTESE 135
VEK ++EV++ E
Sbjct 222 VEKERDKEVSDDE 234
> ath:AT5G56030 HSP81-2 (HEAT SHOCK PROTEIN 81-2); ATP binding;
K04079 molecular chaperone HtpG
Length=699
Score = 123 bits (309), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 55/64 (85%), Positives = 61/64 (95%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEFKA+LFVPKRAPFDLF+T+KK NNIKLYVRR FIMD+CEDIIPE+L FVKG
Sbjct 299 HFSVEGQLEFKAILFVPKRAPFDLFDTKKKPNNIKLYVRRVFIMDNCEDIIPEYLGFVKG 358
Query 61 VVDS 64
+VDS
Sbjct 359 IVDS 362
Score = 99.4 bits (246), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 48/66 (72%), Positives = 56/66 (84%), Gaps = 0/66 (0%)
Query 61 VVDSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIE 120
V +S AGGSFTV +D E LGRGT+++L+LKEDQ EYLEERRLKDLVKKHSEFIS+PI
Sbjct 148 VWESQAGGSFTVTRDTSGETLGRGTKMVLYLKEDQLEYLEERRLKDLVKKHSEFISYPIS 207
Query 121 LAVEKT 126
L +EKT
Sbjct 208 LWIEKT 213
> ath:AT5G56010 HSP81-3; ATP binding / unfolded protein binding;
K04079 molecular chaperone HtpG
Length=699
Score = 123 bits (309), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 55/64 (85%), Positives = 61/64 (95%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEFKA+LFVPKRAPFDLF+T+KK NNIKLYVRR FIMD+CEDIIPE+L FVKG
Sbjct 299 HFSVEGQLEFKAILFVPKRAPFDLFDTKKKPNNIKLYVRRVFIMDNCEDIIPEYLGFVKG 358
Query 61 VVDS 64
+VDS
Sbjct 359 IVDS 362
Score = 99.0 bits (245), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 47/66 (71%), Positives = 56/66 (84%), Gaps = 0/66 (0%)
Query 61 VVDSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIE 120
V +S AGGSFTV +D E LGRGT+++L+LKEDQ EY+EERRLKDLVKKHSEFIS+PI
Sbjct 148 VWESQAGGSFTVTRDTSGEALGRGTKMVLYLKEDQMEYIEERRLKDLVKKHSEFISYPIS 207
Query 121 LAVEKT 126
L +EKT
Sbjct 208 LWIEKT 213
> ath:AT5G52640 ATHSP90.1 (HEAT SHOCK PROTEIN 90.1); ATP binding
/ unfolded protein binding; K04079 molecular chaperone HtpG
Length=705
Score = 123 bits (308), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 55/64 (85%), Positives = 62/64 (96%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEFKA+LFVPKRAPFDLF+TRKK NNIKLYVRR FIMD+CE++IPE+L+FVKG
Sbjct 305 HFSVEGQLEFKAILFVPKRAPFDLFDTRKKLNNIKLYVRRVFIMDNCEELIPEYLSFVKG 364
Query 61 VVDS 64
VVDS
Sbjct 365 VVDS 368
Score = 96.7 bits (239), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 49/66 (74%), Positives = 54/66 (81%), Gaps = 0/66 (0%)
Query 61 VVDSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIE 120
V +S AGGSFTV +D EPLGRGT+I L LK+DQ EYLEERRLKDLVKKHSEFIS+PI
Sbjct 153 VWESQAGGSFTVTRDVDGEPLGRGTKITLFLKDDQLEYLEERRLKDLVKKHSEFISYPIY 212
Query 121 LAVEKT 126
L EKT
Sbjct 213 LWTEKT 218
> hsa:3320 HSP90AA1, FLJ31884, HSP86, HSP89A, HSP90A, HSP90N,
HSPC1, HSPCA, HSPCAL1, HSPCAL4, HSPN, Hsp89, Hsp90, LAP2; heat
shock protein 90kDa alpha (cytosolic), class A member 1;
K04079 molecular chaperone HtpG
Length=854
Score = 123 bits (308), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 53/64 (82%), Positives = 62/64 (96%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEF+ALLFVP+RAPFDLFE RKK+NNIKLYVRR FIMD+CE++IPE+LNF++G
Sbjct 450 HFSVEGQLEFRALLFVPRRAPFDLFENRKKKNNIKLYVRRVFIMDNCEELIPEYLNFIRG 509
Query 61 VVDS 64
VVDS
Sbjct 510 VVDS 513
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 46/70 (65%), Positives = 60/70 (85%), Gaps = 1/70 (1%)
Query 63 DSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELA 122
+S+AGGSFTV+ D EP+GRGT++ILHLKEDQ EYLEERR+K++VKKHS+FI +PI L
Sbjct 285 ESSAGGSFTVRTDTG-EPMGRGTKVILHLKEDQTEYLEERRIKEIVKKHSQFIGYPITLF 343
Query 123 VEKTHEREVT 132
VEK ++EV+
Sbjct 344 VEKERDKEVS 353
> ath:AT5G56000 heat shock protein 81-4 (HSP81-4); K04079 molecular
chaperone HtpG
Length=699
Score = 122 bits (306), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 54/64 (84%), Positives = 61/64 (95%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEFKA+LFVPKRAPFDLF+T+KK NNIKLYVRR FIMD+CEDIIP++L FVKG
Sbjct 299 HFSVEGQLEFKAILFVPKRAPFDLFDTKKKPNNIKLYVRRVFIMDNCEDIIPDYLGFVKG 358
Query 61 VVDS 64
+VDS
Sbjct 359 IVDS 362
Score = 99.8 bits (247), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 48/66 (72%), Positives = 56/66 (84%), Gaps = 0/66 (0%)
Query 61 VVDSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIE 120
V +S AGGSFTV +D E LGRGT++IL+LKEDQ EY+EERRLKDLVKKHSEFIS+PI
Sbjct 148 VWESQAGGSFTVTRDTSGEALGRGTKMILYLKEDQMEYIEERRLKDLVKKHSEFISYPIS 207
Query 121 LAVEKT 126
L +EKT
Sbjct 208 LWIEKT 213
> xla:444024 hsp90aa1.1, MGC82579, hsp86, hsp89, hsp90, hsp90a,
hspc1, hspca, hspn, lap2; heat shock protein 90kDa alpha (cytosolic),
class A member 1, gene 1; K04079 molecular chaperone
HtpG
Length=729
Score = 122 bits (305), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 52/64 (81%), Positives = 62/64 (96%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEF+ALLFVP+RAPFDLFE RKK+NNIKLYVRR FIMD+C+++IPE+LNF++G
Sbjct 325 HFSVEGQLEFRALLFVPRRAPFDLFENRKKKNNIKLYVRRVFIMDNCDELIPEYLNFMRG 384
Query 61 VVDS 64
VVDS
Sbjct 385 VVDS 388
Score = 95.9 bits (237), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 44/70 (62%), Positives = 60/70 (85%), Gaps = 1/70 (1%)
Query 63 DSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELA 122
+S+AGGSFTV+ D+ EPLGRGT++ILHLKEDQ EY EE+R+K++VKKHS+FI +PI L
Sbjct 164 ESSAGGSFTVRVDNS-EPLGRGTKVILHLKEDQSEYFEEKRIKEIVKKHSQFIGYPITLF 222
Query 123 VEKTHEREVT 132
VEK ++E++
Sbjct 223 VEKERDKEIS 232
> dre:30591 hsp90a.1, fb17b01, hsp90, hsp90a, hsp90alpha, wu:fb17b01,
zgc:86652; heat shock protein 90-alpha 1; K04079 molecular
chaperone HtpG
Length=726
Score = 122 bits (305), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 53/64 (82%), Positives = 61/64 (95%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEF+ALLFVP+RA FDLFE +KKRNNIKLYVRR FIMD+CE++IPE+LNF+KG
Sbjct 322 HFSVEGQLEFRALLFVPRRAAFDLFENKKKRNNIKLYVRRVFIMDNCEELIPEYLNFIKG 381
Query 61 VVDS 64
VVDS
Sbjct 382 VVDS 385
Score = 93.2 bits (230), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 45/71 (63%), Positives = 59/71 (83%), Gaps = 1/71 (1%)
Query 61 VVDSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIE 120
+ +SAAGGSFTV K D E +GRGT++ILHLKEDQ EY+EE+R+K++VKKHS+FI +PI
Sbjct 159 IWESAAGGSFTV-KPDFGESIGRGTKVILHLKEDQSEYVEEKRIKEVVKKHSQFIGYPIT 217
Query 121 LAVEKTHEREV 131
L +EK E+EV
Sbjct 218 LYIEKQREKEV 228
> sce:YPL240C HSP82, HSP90; Hsp82p; K04079 molecular chaperone
HtpG
Length=709
Score = 121 bits (303), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 52/64 (81%), Positives = 61/64 (95%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEF+A+LF+PKRAPFDLFE++KK+NNIKLYVRR FI D+ ED+IPEWL+FVKG
Sbjct 308 HFSVEGQLEFRAILFIPKRAPFDLFESKKKKNNIKLYVRRVFITDEAEDLIPEWLSFVKG 367
Query 61 VVDS 64
VVDS
Sbjct 368 VVDS 371
Score = 82.0 bits (201), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 39/71 (54%), Positives = 56/71 (78%), Gaps = 0/71 (0%)
Query 61 VVDSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIE 120
+ +S AGGSFTV D+ E +GRGT + L LK+DQ EYLEE+R+K+++K+HSEF+++PI+
Sbjct 147 IWESNAGGSFTVTLDEVNERIGRGTILRLFLKDDQLEYLEEKRIKEVIKRHSEFVAYPIQ 206
Query 121 LAVEKTHEREV 131
L V K E+EV
Sbjct 207 LVVTKEVEKEV 217
> sce:YMR186W HSC82, HSP90; Cytoplasmic chaperone of the Hsp90
family, redundant in function and nearly identical with Hsp82p,
and together they are essential; expressed constitutively
at 10-fold higher basal levels than HSP82 and induced 2-3
fold by heat shock; K04079 molecular chaperone HtpG
Length=705
Score = 121 bits (303), Expect = 9e-28, Method: Compositional matrix adjust.
Identities = 52/64 (81%), Positives = 61/64 (95%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEF+A+LF+PKRAPFDLFE++KK+NNIKLYVRR FI D+ ED+IPEWL+FVKG
Sbjct 304 HFSVEGQLEFRAILFIPKRAPFDLFESKKKKNNIKLYVRRVFITDEAEDLIPEWLSFVKG 363
Query 61 VVDS 64
VVDS
Sbjct 364 VVDS 367
Score = 81.6 bits (200), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 39/71 (54%), Positives = 56/71 (78%), Gaps = 0/71 (0%)
Query 61 VVDSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIE 120
+ +S AGGSFTV D+ E +GRGT + L LK+DQ EYLEE+R+K+++K+HSEF+++PI+
Sbjct 147 IWESNAGGSFTVTLDEVNERIGRGTVLRLFLKDDQLEYLEEKRIKEVIKRHSEFVAYPIQ 206
Query 121 LAVEKTHEREV 131
L V K E+EV
Sbjct 207 LLVTKEVEKEV 217
> cel:C47E8.5 daf-21; abnormal DAuer Formation family member (daf-21);
K04079 molecular chaperone HtpG
Length=702
Score = 116 bits (291), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 49/64 (76%), Positives = 61/64 (95%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEF+ALLFVP+RAPFDLFE +K +N+IKLYVRR FIM++CE+++PE+LNF+KG
Sbjct 299 HFSVEGQLEFRALLFVPQRAPFDLFENKKSKNSIKLYVRRVFIMENCEELMPEYLNFIKG 358
Query 61 VVDS 64
VVDS
Sbjct 359 VVDS 362
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 32/59 (54%), Positives = 50/59 (84%), Gaps = 1/59 (1%)
Query 63 DSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIEL 121
+S+AGGSF V+ + E + RGT+I++H+KEDQ ++LEER++K++VKKHS+FI +PI+L
Sbjct 151 ESSAGGSFVVRPFNDPE-VTRGTKIVMHIKEDQIDFLEERKIKEIVKKHSQFIGYPIKL 208
> dre:565155 hsp90a.2, cb820, hsp90a2; heat shock protein 90-alpha
2; K04079 molecular chaperone HtpG
Length=734
Score = 105 bits (263), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 52/64 (81%), Positives = 62/64 (96%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEF+ALLFVP+RAPFDLFE +KK+NNIKLYVRR FIMD+C+++IPE+LNF+KG
Sbjct 330 HFSVEGQLEFRALLFVPRRAPFDLFENKKKKNNIKLYVRRVFIMDNCDELIPEYLNFIKG 389
Query 61 VVDS 64
VVDS
Sbjct 390 VVDS 393
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 48/92 (52%), Positives = 72/92 (78%), Gaps = 1/92 (1%)
Query 41 AFIMDDCEDIIPEWLNFVKGVVDSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLE 100
A+++ + +I + L+ + +S+AGGSFTV+ D+ EP+GRGT++ILHLKEDQ EY+E
Sbjct 139 AYLVAEKVTVITKHLDDEQYAWESSAGGSFTVKVDNS-EPIGRGTKVILHLKEDQTEYIE 197
Query 101 ERRLKDLVKKHSEFISFPIELAVEKTHEREVT 132
ERR+K++VKKHS+FI +PI L VEK ++EV+
Sbjct 198 ERRIKEIVKKHSQFIGYPITLFVEKERDKEVS 229
> dre:30573 hsp90ab1, hsp90b, hsp90beta, wu:fa29f01, wu:fa91e11,
wu:fd59e11, wu:gcd22h07; heat shock protein 90, alpha (cytosolic),
class B member 1; K04079 molecular chaperone HtpG
Length=725
Score = 105 bits (262), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 51/64 (79%), Positives = 62/64 (96%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEF+ALLF+P+RAPFDLFE +KK+NNIKLYVRR FIMD+CE++IPE+LNF++G
Sbjct 319 HFSVEGQLEFRALLFIPRRAPFDLFENKKKKNNIKLYVRRVFIMDNCEELIPEYLNFIRG 378
Query 61 VVDS 64
VVDS
Sbjct 379 VVDS 382
Score = 93.6 bits (231), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 44/70 (62%), Positives = 60/70 (85%), Gaps = 1/70 (1%)
Query 63 DSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELA 122
+S+AGGSFTV K D EP+GRGT++ILHLKEDQ EY+EE+R+K++VKKHS+FI +PI L
Sbjct 157 ESSAGGSFTV-KVDHGEPIGRGTKVILHLKEDQTEYIEEKRVKEVVKKHSQFIGYPITLY 215
Query 123 VEKTHEREVT 132
VEK ++E++
Sbjct 216 VEKERDKEIS 225
> xla:446459 hsp90ab1, hsp90b, hsp90beta; heat shock protein 90kDa
alpha (cytosolic), class B member 1; K04079 molecular chaperone
HtpG
Length=722
Score = 104 bits (259), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 51/64 (79%), Positives = 61/64 (95%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEF+ALLF+P+RAPFDLFE +KK+NNIKLYVRR FIMD C+++IPE+LNFV+G
Sbjct 318 HFSVEGQLEFRALLFIPRRAPFDLFENKKKKNNIKLYVRRVFIMDSCDELIPEYLNFVRG 377
Query 61 VVDS 64
VVDS
Sbjct 378 VVDS 381
Score = 94.4 bits (233), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 45/71 (63%), Positives = 60/71 (84%), Gaps = 1/71 (1%)
Query 63 DSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELA 122
+S+AGGSFTV K D EP+GRGT++ILHLKEDQ EYLEE+R+K+ VKKHS+FI +PI L
Sbjct 158 ESSAGGSFTV-KVDTGEPIGRGTKVILHLKEDQTEYLEEKRVKETVKKHSQFIGYPITLY 216
Query 123 VEKTHEREVTE 133
+EK E+E+++
Sbjct 217 LEKEREKEISD 227
> mmu:15516 Hsp90ab1, 90kDa, AL022974, C81438, Hsp84, Hsp84-1,
Hsp90, Hspcb, MGC115780; heat shock protein 90 alpha (cytosolic),
class B member 1; K04079 molecular chaperone HtpG
Length=724
Score = 103 bits (258), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 50/64 (78%), Positives = 61/64 (95%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEF+ALLF+P+RAPFDLFE +KK+NNIKLYVRR FIMD C+++IPE+LNF++G
Sbjct 320 HFSVEGQLEFRALLFIPRRAPFDLFENKKKKNNIKLYVRRVFIMDSCDELIPEYLNFIRG 379
Query 61 VVDS 64
VVDS
Sbjct 380 VVDS 383
Score = 96.3 bits (238), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 45/70 (64%), Positives = 60/70 (85%), Gaps = 1/70 (1%)
Query 63 DSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELA 122
+S+AGGSFTV+ D EP+GRGT++ILHLKEDQ EYLEERR+K++VKKHS+FI +PI L
Sbjct 158 ESSAGGSFTVRADHG-EPIGRGTKVILHLKEDQTEYLEERRVKEVVKKHSQFIGYPITLY 216
Query 123 VEKTHEREVT 132
+EK E+E++
Sbjct 217 LEKEREKEIS 226
> hsa:3326 HSP90AB1, D6S182, FLJ26984, HSP90-BETA, HSP90B, HSPC2,
HSPCB; heat shock protein 90kDa alpha (cytosolic), class
B member 1; K04079 molecular chaperone HtpG
Length=724
Score = 103 bits (258), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 50/64 (78%), Positives = 61/64 (95%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HFSVEGQLEF+ALLF+P+RAPFDLFE +KK+NNIKLYVRR FIMD C+++IPE+LNF++G
Sbjct 320 HFSVEGQLEFRALLFIPRRAPFDLFENKKKKNNIKLYVRRVFIMDSCDELIPEYLNFIRG 379
Query 61 VVDS 64
VVDS
Sbjct 380 VVDS 383
Score = 96.3 bits (238), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 45/70 (64%), Positives = 60/70 (85%), Gaps = 1/70 (1%)
Query 63 DSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELA 122
+S+AGGSFTV+ D EP+GRGT++ILHLKEDQ EYLEERR+K++VKKHS+FI +PI L
Sbjct 158 ESSAGGSFTVRADHG-EPIGRGTKVILHLKEDQTEYLEERRVKEVVKKHSQFIGYPITLY 216
Query 123 VEKTHEREVT 132
+EK E+E++
Sbjct 217 LEKEREKEIS 226
> cpv:cgd7_3670 heat shock protein 90 (Hsp90), signal peptide
plus ER retention motif
Length=787
Score = 89.7 bits (221), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 39/65 (60%), Positives = 50/65 (76%), Gaps = 1/65 (1%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFET-RKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVK 59
HFS EG++EFK+LLF+P PFD+F+T K NIK YVRR I D ED++P++LNF+K
Sbjct 355 HFSAEGEIEFKSLLFIPSHPPFDMFDTYMGKSGNIKFYVRRVLITDHIEDLLPKYLNFIK 414
Query 60 GVVDS 64
GVVDS
Sbjct 415 GVVDS 419
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 53/83 (63%), Gaps = 1/83 (1%)
Query 61 VVDSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIE 120
V +S+A GSF V D + + RGT I+L LKED E++ +LKDLV ++S+FI+FPI
Sbjct 230 VWESSADGSFRVSLDPRGNTIKRGTTIVLSLKEDATEFMNFSKLKDLVLRYSQFINFPIY 289
Query 121 L-AVEKTHEREVTESEDEEEKKA 142
+ E ++ E ES +++E K
Sbjct 290 IYNPEGVNKSEKDESGEKKESKG 312
> hsa:7184 HSP90B1, ECGP, GP96, GRP94, TRA1; heat shock protein
90kDa beta (Grp94), member 1; K09487 heat shock protein 90kDa
beta
Length=803
Score = 82.4 bits (202), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 39/66 (59%), Positives = 50/66 (75%), Gaps = 2/66 (3%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFE--TRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFV 58
HF+ EG++ FK++LFVP AP LF+ KK + IKLYVRR FI DD D++P++LNFV
Sbjct 374 HFTAEGEVTFKSILFVPTSAPRGLFDEYGSKKSDYIKLYVRRVFITDDFHDMMPKYLNFV 433
Query 59 KGVVDS 64
KGVVDS
Sbjct 434 KGVVDS 439
Score = 55.1 bits (131), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 27/50 (54%), Positives = 36/50 (72%), Gaps = 0/50 (0%)
Query 70 FTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPI 119
F+V D + LGRGT I L LKE+ +YLE +K+LVKK+S+FI+FPI
Sbjct 230 FSVIADPRGNTLGRGTTITLVLKEEASDYLELDTIKNLVKKYSQFINFPI 279
> mmu:22027 Hsp90b1, ERp99, GRP94, TA-3, Targ2, Tra-1, Tra1, endoplasmin,
gp96; heat shock protein 90, beta (Grp94), member
1; K09487 heat shock protein 90kDa beta
Length=802
Score = 82.4 bits (202), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 39/66 (59%), Positives = 50/66 (75%), Gaps = 2/66 (3%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFE--TRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFV 58
HF+ EG++ FK++LFVP AP LF+ KK + IKLYVRR FI DD D++P++LNFV
Sbjct 374 HFTAEGEVTFKSILFVPTSAPRGLFDEYGSKKSDYIKLYVRRVFITDDFHDMMPKYLNFV 433
Query 59 KGVVDS 64
KGVVDS
Sbjct 434 KGVVDS 439
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 29/61 (47%), Positives = 40/61 (65%), Gaps = 0/61 (0%)
Query 70 FTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELAVEKTHER 129
F+V D + LGRGT I L LKE+ +YLE +K+LV+K+S+FI+FPI + KT
Sbjct 230 FSVIADPRGNTLGRGTTITLVLKEEASDYLELDTIKNLVRKYSQFINFPIYVWSSKTETV 289
Query 130 E 130
E
Sbjct 290 E 290
> tgo:TGME49_044560 heat shock protein 90, putative (EC:2.7.13.3);
K09487 heat shock protein 90kDa beta
Length=847
Score = 82.4 bits (202), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 35/65 (53%), Positives = 52/65 (80%), Gaps = 1/65 (1%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETR-KKRNNIKLYVRRAFIMDDCEDIIPEWLNFVK 59
HFS EG++EFKALL++PKRAP D++ K+ ++K+YVRR + D +D++P++L+FVK
Sbjct 391 HFSAEGEVEFKALLYIPKRAPSDIYSNYFDKQTSVKVYVRRVLVADQFDDLLPKYLHFVK 450
Query 60 GVVDS 64
GVVDS
Sbjct 451 GVVDS 455
Score = 68.9 bits (167), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 42/130 (32%), Positives = 69/130 (53%), Gaps = 6/130 (4%)
Query 16 VPKRAPFDLFETRKKRNNIKLYVR------RAFIMDDCEDIIPEWLNFVKGVVDSAAGGS 69
V K + E + N++ L + AF++ D ++ + + + + +S+A
Sbjct 178 VAKSGTSNFLEAMAQGNDVNLIGQFGVGFYSAFLVADKVTVVSKNVEDDQHIWESSADAK 237
Query 70 FTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELAVEKTHER 129
F V KD + LGRGT + LHLKED E+L E +LKDL + S+F+S+PI + +T
Sbjct 238 FHVAKDPRGNTLGRGTCVTLHLKEDATEFLNEWKLKDLTTRFSQFMSYPIYVRTSRTVTE 297
Query 130 EVTESEDEEE 139
EV ++E E
Sbjct 298 EVPIEDEEAE 307
> cel:T05E11.3 hypothetical protein; K09487 heat shock protein
90kDa beta
Length=760
Score = 82.0 bits (201), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 34/65 (52%), Positives = 52/65 (80%), Gaps = 1/65 (1%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKR-NNIKLYVRRAFIMDDCEDIIPEWLNFVK 59
HFS EG++ F+++L+VPK++P D+F+ K NIKLYVRR FI DD D++P++L+F++
Sbjct 353 HFSAEGEVSFRSILYVPKKSPNDMFQNYGKVIENIKLYVRRVFITDDFADMLPKYLSFIR 412
Query 60 GVVDS 64
G+VDS
Sbjct 413 GIVDS 417
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 29/60 (48%), Positives = 40/60 (66%), Gaps = 0/60 (0%)
Query 67 GGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELAVEKT 126
SFT+ KD + L RGT+I L+LKE+ ++LE LK+LV K+S+FI+F I L KT
Sbjct 215 SASFTISKDPRGNTLKRGTQITLYLKEEAADFLEPDTLKNLVHKYSQFINFDIFLWQSKT 274
> ath:AT4G24190 SHD; SHD (SHEPHERD); ATP binding / unfolded protein
binding
Length=823
Score = 80.9 bits (198), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 35/66 (53%), Positives = 53/66 (80%), Gaps = 2/66 (3%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRN--NIKLYVRRAFIMDDCEDIIPEWLNFV 58
HF+ EG +EFKA+L+VP +AP DL+E+ N N+KLYVRR FI D+ ++++P++L+F+
Sbjct 381 HFNAEGDVEFKAVLYVPPKAPHDLYESYYNSNKANLKLYVRRVFISDEFDELLPKYLSFL 440
Query 59 KGVVDS 64
KG+VDS
Sbjct 441 KGLVDS 446
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 41/85 (48%), Positives = 59/85 (69%), Gaps = 0/85 (0%)
Query 41 AFIMDDCEDIIPEWLNFVKGVVDSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLE 100
A+++ D ++I + + + V +S A G F V +D EPLGRGT I LHL+++ GEYLE
Sbjct 202 AYLVADYIEVISKHNDDSQYVWESKANGKFAVSEDTWNEPLGRGTEIRLHLRDEAGEYLE 261
Query 101 ERRLKDLVKKHSEFISFPIELAVEK 125
E +LK+LVK++SEFI+FPI L K
Sbjct 262 ESKLKELVKRYSEFINFPISLWASK 286
> dre:386590 hsp90b1, GP96, GRP94, fb61d09, tra-1, tra1, wu:fb61d09,
wu:fq25g01; heat shock protein 90, beta (grp94), member
1; K09487 heat shock protein 90kDa beta
Length=793
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 37/66 (56%), Positives = 50/66 (75%), Gaps = 2/66 (3%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFE--TRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFV 58
HF+ EG++ FK++LFVP AP LF+ KK + IKL+VRR FI DD D++P++LNF+
Sbjct 374 HFTAEGEVTFKSILFVPASAPRGLFDEYGTKKNDFIKLFVRRVFITDDFHDMMPKYLNFI 433
Query 59 KGVVDS 64
KGVVDS
Sbjct 434 KGVVDS 439
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 29/64 (45%), Positives = 42/64 (65%), Gaps = 0/64 (0%)
Query 67 GGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELAVEKT 126
F+V +D + + LGRGT I L +KE+ +YLE +K+LVKK+S+FI+FPI + KT
Sbjct 227 SNQFSVIEDPRGDTLGRGTTITLVMKEEASDYLELETIKNLVKKYSQFINFPIYVWSSKT 286
Query 127 HERE 130
E
Sbjct 287 ETVE 290
> xla:399408 hsp90b1, ecgp, gp96, grp94, tra1; heat shock protein
90kDa beta (Grp94), member 1
Length=804
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 37/66 (56%), Positives = 50/66 (75%), Gaps = 2/66 (3%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFE--TRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFV 58
HF+ EG++ FK++LF+P AP LF+ KK + IKL+VRR FI DD D++P++LNFV
Sbjct 373 HFTAEGEVTFKSILFIPSSAPRGLFDEYGSKKSDFIKLFVRRVFITDDFHDMMPKYLNFV 432
Query 59 KGVVDS 64
KGVVDS
Sbjct 433 KGVVDS 438
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 27/57 (47%), Positives = 39/57 (68%), Gaps = 0/57 (0%)
Query 70 FTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELAVEKT 126
F V D + + LGRG+ I L LKE+ +YLE +K+LV+K+S+FI+FPI + KT
Sbjct 230 FFVTDDPRGDTLGRGSTITLVLKEEATDYLELETVKNLVRKYSQFINFPIYVWSSKT 286
> xla:398753 hypothetical protein MGC68448; K09487 heat shock
protein 90kDa beta
Length=805
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 37/66 (56%), Positives = 50/66 (75%), Gaps = 2/66 (3%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFE--TRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFV 58
HF+ EG++ FK++LF+P AP LF+ KK + IKL+VRR FI DD D++P++LNFV
Sbjct 373 HFTAEGEVTFKSILFIPSTAPRGLFDEYGSKKIDFIKLFVRRVFITDDFNDMMPKYLNFV 432
Query 59 KGVVDS 64
KGVVDS
Sbjct 433 KGVVDS 438
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/61 (45%), Positives = 40/61 (65%), Gaps = 0/61 (0%)
Query 70 FTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELAVEKTHER 129
F V D + + LGRGT I L LKE+ +YLE +K+LV+K+S+F++FPI + KT
Sbjct 230 FFVTDDPRGDTLGRGTTITLVLKEEATDYLELETIKNLVRKYSQFMNFPIYVWSSKTETV 289
Query 130 E 130
E
Sbjct 290 E 290
> eco:b0473 htpG, ECK0467, JW0462; molecular chaperone HSP90 family;
K04079 molecular chaperone HtpG
Length=624
Score = 74.7 bits (182), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 30/65 (46%), Positives = 49/65 (75%), Gaps = 1/65 (1%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
H VEG+ E+ +LL++P +AP+D++ R ++ +KLYV+R FIMDD E +P +L FV+G
Sbjct 265 HNRVEGKQEYTSLLYIPSQAPWDMW-NRDHKHGLKLYVQRVFIMDDAEQFMPNYLRFVRG 323
Query 61 VVDSA 65
++DS+
Sbjct 324 LIDSS 328
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/68 (36%), Positives = 44/68 (64%), Gaps = 5/68 (7%)
Query 63 DSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELA 122
+SA G +TV K + RGT I LHL+E + E+L++ R++ ++ K+S+ I+ P+E
Sbjct 155 ESAGEGEYTVADITKED---RGTEITLHLREGEDEFLDDWRVRSIISKYSDHIALPVE-- 209
Query 123 VEKTHERE 130
+EK E++
Sbjct 210 IEKREEKD 217
> bbo:BBOV_III007380 17.m07646; heat shock protein 90
Length=795
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 33/66 (50%), Positives = 42/66 (63%), Gaps = 2/66 (3%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETR--KKRNNIKLYVRRAFIMDDCEDIIPEWLNFV 58
HF VEGQ+EF LLFVP P++L + I+LYV+R FI D + +P WL FV
Sbjct 406 HFKVEGQVEFSCLLFVPGSLPWELSRNMFDDQSRGIRLYVKRVFINDKFSEAVPRWLTFV 465
Query 59 KGVVDS 64
+GVVDS
Sbjct 466 RGVVDS 471
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 34/82 (41%), Positives = 55/82 (67%), Gaps = 5/82 (6%)
Query 64 SAAGGSFTVQK--DDKYEP--LGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPI 119
S G+F+V + DD+ + + GTRI+LH+K + +YLE+ ++K+L++K+SEF+ FPI
Sbjct 270 SETNGTFSVAQVNDDELQKGFMKCGTRIVLHIKPECDDYLEDYKIKELLRKYSEFVRFPI 329
Query 120 ELAVEKT-HEREVTESEDEEEK 140
++ VEK +ER ES E K
Sbjct 330 QVWVEKVEYERVPDESTAVEGK 351
> tpv:TP04_0646 heat shock protein 90
Length=913
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 34/66 (51%), Positives = 42/66 (63%), Gaps = 2/66 (3%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETR--KKRNNIKLYVRRAFIMDDCEDIIPEWLNFV 58
HF VEGQ+EF LLFVP P++L + I+LYV+R FI D + IP WL FV
Sbjct 415 HFKVEGQVEFTCLLFVPGTLPWELSRNMFDDESRGIRLYVKRVFINDKFSESIPRWLTFV 474
Query 59 KGVVDS 64
+GVVDS
Sbjct 475 RGVVDS 480
Score = 58.5 bits (140), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 29/68 (42%), Positives = 49/68 (72%), Gaps = 8/68 (11%)
Query 64 SAAGGSFTVQK------DDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISF 117
S + G++T+ + +DK+ + GTRI+LHLK + +YLE+ +LK+L++K+SEFI F
Sbjct 279 SDSNGTYTIGRVENQELNDKF--MKSGTRIVLHLKPECDDYLEDYKLKELLRKYSEFIRF 336
Query 118 PIELAVEK 125
PI++ VE+
Sbjct 337 PIQVWVER 344
> pfa:PFL1070c endoplasmin homolog precursor, putative
Length=821
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 29/64 (45%), Positives = 46/64 (71%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HF EG++EFK L+++P +AP + K+N++KLYVRR + D+ + +P +++FVKG
Sbjct 367 HFFAEGEIEFKCLIYIPSKAPSMNDQLYSKQNSLKLYVRRVLVADEFVEFLPRYMSFVKG 426
Query 61 VVDS 64
VVDS
Sbjct 427 VVDS 430
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 33/71 (46%), Positives = 44/71 (61%), Gaps = 0/71 (0%)
Query 61 VVDSAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIE 120
+ +S A FT+ KD + L RGTRI LHLKED L +++L DL+ K+S+FI FPI
Sbjct 217 IWESTADAKFTIYKDPRGATLKRGTRISLHLKEDATNLLNDKKLMDLISKYSQFIQFPIY 276
Query 121 LAVEKTHEREV 131
L E + EV
Sbjct 277 LLHENVYTEEV 287
> pfa:PF14_0417 HSP90
Length=927
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 42/99 (42%), Positives = 58/99 (58%), Gaps = 20/99 (20%)
Query 32 NNIKLYVRRAFIMDDCEDIIPEWLNFVKGVVDSAAGGSFTVQKDDKYEP-----LGRGTR 86
N +++Y ++ ED I W S GSF+V + KY+ G GT+
Sbjct 265 NRVEVYTKK-------EDQIYRW--------SSDLKGSFSVNEIKKYDQEYDDIKGSGTK 309
Query 87 IILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELAVEK 125
IILHLKE+ EYLE+ +LK+L+KK+SEFI FPIE+ EK
Sbjct 310 IILHLKEECDEYLEDYKLKELIKKYSEFIKFPIEIWSEK 348
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 28/66 (42%), Positives = 45/66 (68%), Gaps = 2/66 (3%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETR--KKRNNIKLYVRRAFIMDDCEDIIPEWLNFV 58
HF+VEGQ+ F ++L++P P++L + ++ I+LYV+R FI D + IP WL F+
Sbjct 420 HFNVEGQISFNSILYIPGSLPWELSKNMFDEESRGIRLYVKRVFINDKFSESIPRWLTFL 479
Query 59 KGVVDS 64
+G+VDS
Sbjct 480 RGIVDS 485
> ath:AT2G04030 CR88; CR88; ATP binding
Length=780
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 48/66 (72%), Gaps = 2/66 (3%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFE-TRKKRNNIKLYVRRAFIMDDCE-DIIPEWLNFV 58
HF+ EG++EF+++L++P P + + T K NI+LYV+R FI DD + ++ P +L+FV
Sbjct 367 HFTTEGEVEFRSILYIPGMGPLNNEDVTNPKTKNIRLYVKRVFISDDFDGELFPRYLSFV 426
Query 59 KGVVDS 64
KGVVDS
Sbjct 427 KGVVDS 432
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/56 (48%), Positives = 38/56 (67%), Gaps = 1/56 (1%)
Query 81 LGRGTRIILHLKED-QGEYLEERRLKDLVKKHSEFISFPIELAVEKTHEREVTESE 135
L RGT+I L+L+ED + E+ E R+K+LVK +S+F+ FPI EK+ EV E E
Sbjct 247 LRRGTQITLYLREDDKYEFAESTRIKNLVKNYSQFVGFPIYTWQEKSRTIEVEEDE 302
> tgo:TGME49_110430 heat shock protein 90, putative (EC:3.2.1.3
2.7.13.3)
Length=1100
Score = 66.2 bits (160), Expect = 3e-11, Method: Composition-based stats.
Identities = 31/66 (46%), Positives = 43/66 (65%), Gaps = 2/66 (3%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETR--KKRNNIKLYVRRAFIMDDCEDIIPEWLNFV 58
H VEGQ++F ALLF+P P++L ++ I+LYV+R FI D D +P WL F+
Sbjct 646 HMKVEGQVDFNALLFIPGALPWELARNMFDEESRGIRLYVKRVFINDKFADAVPRWLTFI 705
Query 59 KGVVDS 64
+GVVDS
Sbjct 706 RGVVDS 711
Score = 57.8 bits (138), Expect = 1e-08, Method: Composition-based stats.
Identities = 25/54 (46%), Positives = 38/54 (70%), Gaps = 0/54 (0%)
Query 84 GTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELAVEKTHEREVTESEDE 137
GTR++LHLKED +YLE+ +LK+L++K+SEFI PI + E+ V E ++
Sbjct 484 GTRVVLHLKEDSDDYLEDYKLKELMRKYSEFIQLPIHIWSERIEYERVPEGSEQ 537
> ath:AT3G07770 ATP binding
Length=799
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 32/68 (47%), Positives = 49/68 (72%), Gaps = 6/68 (8%)
Query 1 HFSVEGQLEFKALLFVPKRAPF---DLFETRKKRNNIKLYVRRAFIMDDCE-DIIPEWLN 56
HF+ EG++EF+++L+VP +P D+ + K NI+LYV+R FI DD + ++ P +L+
Sbjct 390 HFTTEGEVEFRSILYVPPVSPSGKDDIVNQKTK--NIRLYVKRVFISDDFDGELFPRYLS 447
Query 57 FVKGVVDS 64
FVKGVVDS
Sbjct 448 FVKGVVDS 455
Score = 62.0 bits (149), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 33/78 (42%), Positives = 48/78 (61%), Gaps = 1/78 (1%)
Query 65 AAGGSFTVQKD-DKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELAV 123
A SFT+Q+D D + RGTRI LHLK++ + + R++ LVK +S+F+SFPI
Sbjct 248 ANSSSFTIQEDTDPQSLIPRGTRITLHLKQEAKNFADPERIQKLVKNYSQFVSFPIYTWQ 307
Query 124 EKTHEREVTESEDEEEKK 141
EK + +EV +D E K
Sbjct 308 EKGYTKEVEVEDDPTETK 325
> tpv:TP01_0934 heat shock protein 90
Length=1009
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 28/66 (42%), Positives = 40/66 (60%), Gaps = 0/66 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HF+ EG ++FKALL++P P F + +N+KLY RR + + D IP +L V G
Sbjct 337 HFTAEGDVDFKALLYIPSSPPAMYFSSESVGHNVKLYSRRVLVSQEMRDFIPRYLFSVYG 396
Query 61 VVDSAA 66
VVDS +
Sbjct 397 VVDSDS 402
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 23/58 (39%), Positives = 37/58 (63%), Gaps = 0/58 (0%)
Query 64 SAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIEL 121
S+A S+ + +D GT I L L+ED +YL+ L++LVKK+S+F+ +PI+L
Sbjct 227 SSAANSYELYEDTDNSLGDHGTLITLELREDATDYLKTDVLENLVKKYSQFVKYPIQL 284
> bbo:BBOV_IV008400 23.m06066; heat shock protein 90; K09487 heat
shock protein 90kDa beta
Length=795
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 39/64 (60%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
HF EG ++F+ALL++P+R F+ +++K+Y RR + D + +P +L + G
Sbjct 346 HFVAEGDIDFRALLYIPERPKSAYFDNEDVGHHVKIYARRVLVSDSLPNFLPRYLYSLHG 405
Query 61 VVDS 64
VVDS
Sbjct 406 VVDS 409
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 26/57 (45%), Positives = 39/57 (68%), Gaps = 1/57 (1%)
Query 64 SAAGGSFTVQKDDKYEPLG-RGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPI 119
S+A + + +D K LG GT+I L L+ED EYLE ++++L+KKHS+F+ FPI
Sbjct 235 SSADTKYELYEDPKGNTLGEHGTQITLFLREDATEYLEIDKIEELIKKHSQFVRFPI 291
> mmu:68015 Trap1, 2410002K23Rik, HSP75; TNF receptor-associated
protein 1; K09488 TNF receptor-associated protein 1
Length=706
Score = 55.1 bits (131), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 38/64 (59%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
H+ + L +++ +VP+ P +R+ +++ LY R+ I DI+P+WL F++G
Sbjct 332 HYKTDAPLNIRSIFYVPEMKPSMFDVSRELGSSVALYSRKVLIQTKAADILPKWLRFIRG 391
Query 61 VVDS 64
VVDS
Sbjct 392 VVDS 395
Score = 48.1 bits (113), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 23/58 (39%), Positives = 35/58 (60%), Gaps = 3/58 (5%)
Query 64 SAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIEL 121
S G F + + P GT+II+HLK D ++ E R++D+V K+S F+SFP+ L
Sbjct 235 SDGSGVFEIAEASGVRP---GTKIIIHLKSDCKDFASESRVQDVVTKYSNFVSFPLYL 289
> hsa:10131 TRAP1, HSP75, HSP90L; TNF receptor-associated protein
1; K09488 TNF receptor-associated protein 1
Length=704
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 37/64 (57%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
H+ + L +++ +VP P +R+ +++ LY R+ I DI+P+WL F++G
Sbjct 330 HYKTDAPLNIRSIFYVPDMKPSMFDVSRELGSSVALYSRKVLIQTKATDILPKWLRFIRG 389
Query 61 VVDS 64
VVDS
Sbjct 390 VVDS 393
Score = 48.1 bits (113), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 40/70 (57%), Gaps = 13/70 (18%)
Query 53 EWLNFVKGVVDSA-AGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKH 111
+WL+ GV + A A G T GT+II+HLK D E+ E R++D+V K+
Sbjct 230 QWLSDGSGVFEIAEASGVRT------------GTKIIIHLKSDCKEFSSEARVRDVVTKY 277
Query 112 SEFISFPIEL 121
S F+SFP+ L
Sbjct 278 SNFVSFPLYL 287
> dre:571959 trap1, fc85a11, wu:fc85a11; TNF receptor-associated
protein 1; K09488 TNF receptor-associated protein 1
Length=719
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 38/64 (59%), Gaps = 0/64 (0%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKG 60
H+ + L +++ +VP+ P +R+ +++ LY R+ I DI+P+WL F++G
Sbjct 345 HYRADAPLNIRSIFYVPEMKPSMFDVSREMGSSVALYSRKILIQTKATDILPKWLRFLRG 404
Query 61 VVDS 64
VVDS
Sbjct 405 VVDS 408
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 21/39 (53%), Positives = 31/39 (79%), Gaps = 0/39 (0%)
Query 83 RGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIEL 121
+GT+I+LHLK+D E+ E R+K++V K+S F+SFPI L
Sbjct 264 QGTKIVLHLKDDCKEFSSEDRVKEVVTKYSNFVSFPIFL 302
> tgo:TGME49_092920 heat shock protein 90, putative ; K09488 TNF
receptor-associated protein 1
Length=861
Score = 50.4 bits (119), Expect = 2e-06, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 35/62 (56%), Gaps = 1/62 (1%)
Query 2 FSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKGV 61
F + L KA+ ++P+ P LF+ + + L+ RR + DIIP+WL FVKGV
Sbjct 419 FRTDAPLSIKAVFYIPEDPPSRLFQPANEVG-VSLHSRRVLVKKSATDIIPKWLGFVKGV 477
Query 62 VD 63
+D
Sbjct 478 ID 479
Score = 39.7 bits (91), Expect = 0.003, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 31/55 (56%), Gaps = 0/55 (0%)
Query 81 LGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELAVEKTHEREVTESE 135
L RGT+I+ HLK+D E+ +K+ K S F++FPI + E ++T +
Sbjct 331 LKRGTKIVCHLKKDCLEFSNIHHVKECATKFSSFVNFPIYVKEEDGKNTKITSQQ 385
> bbo:BBOV_IV010880 23.m05763; heat shock protein 75; K09488 TNF
receptor-associated protein 1
Length=623
Score = 48.9 bits (115), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 35/62 (56%), Gaps = 1/62 (1%)
Query 2 FSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKGV 61
F + L K+LL++P+ AP LF + + L+ R+ I IIP+WL F+KGV
Sbjct 274 FHSDVPLSIKSLLYIPEDAPSKLFHNTNEVG-VSLHSRKILIQKSATAIIPKWLFFIKGV 332
Query 62 VD 63
+D
Sbjct 333 ID 334
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 41/69 (59%), Gaps = 4/69 (5%)
Query 64 SAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELAV 123
S GSFT+++ + L RGT+I+ HL++D + +K + +K S F++FP+ +
Sbjct 173 SDGTGSFTIRE---VQELPRGTKIVCHLRDDCVVFANTANVKKVAEKFSAFVNFPLYIQ- 228
Query 124 EKTHEREVT 132
EK E E+T
Sbjct 229 EKDAETEIT 237
> pfa:PF11_0188 heat shock protein 90, putative
Length=930
Score = 47.8 bits (112), Expect = 1e-05, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 38/62 (61%), Gaps = 1/62 (1%)
Query 2 FSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVKGV 61
+ + L K++ ++P+ AP LF+ + I LY ++ + + ++IIP+WL FVKGV
Sbjct 495 YKTDAPLSIKSVFYIPEEAPSRLFQ-QSNDIEISLYCKKVLVKKNADNIIPKWLYFVKGV 553
Query 62 VD 63
+D
Sbjct 554 ID 555
Score = 38.1 bits (87), Expect = 0.008, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 39/62 (62%), Gaps = 3/62 (4%)
Query 64 SAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELAV 123
S G+FT+++ D + +GT+II HLK+ E+ + ++ +V+K S FI+FP+ +
Sbjct 255 SYGNGTFTLKEVDN---IPKGTKIICHLKDSCKEFSNIQNVQKIVEKFSSFINFPVYVLK 311
Query 124 EK 125
+K
Sbjct 312 KK 313
> tpv:TP01_0753 heat shock protein 75; K09488 TNF receptor-associated
protein 1
Length=724
Score = 46.6 bits (109), Expect = 3e-05, Method: Composition-based stats.
Identities = 24/65 (36%), Positives = 38/65 (58%), Gaps = 3/65 (4%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDLFETRKKRNNIKLYVR--RAFIMDDCEDIIPEWLNFV 58
+F + L K+L ++P+ AP +F+ + + LY R + I E+IIP+WL FV
Sbjct 334 NFHSDSPLSIKSLFYIPEDAPNRMFQASNELG-VSLYSRYLKVLIKKSAENIIPKWLFFV 392
Query 59 KGVVD 63
KGV+D
Sbjct 393 KGVID 397
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 27/69 (39%), Positives = 41/69 (59%), Gaps = 4/69 (5%)
Query 64 SAAGGSFTVQKDDKYEPLGRGTRIILHLKEDQGEYLEERRLKDLVKKHSEFISFPIELAV 123
S GSFT+++ D L RGT+II +LK+D + +K + +K S FI+FP+ L
Sbjct 234 SDGTGSFTLKEVDN---LPRGTKIICYLKDDSLLFCNSNNVKKVAEKFSSFINFPLFLQ- 289
Query 124 EKTHEREVT 132
EK + E+T
Sbjct 290 EKDKDVEIT 298
> cel:R151.7 hypothetical protein
Length=479
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 37/65 (56%), Gaps = 1/65 (1%)
Query 1 HFSVEGQLEFKALLFVPKRAPFDL-FETRKKRNNIKLYVRRAFIMDDCEDIIPEWLNFVK 59
HF + + ++++++P+ L F ++ + LY RR I D +++IP +L FV
Sbjct 292 HFQTDTPVSLRSVIYIPQTQFNQLTFMAQQTMCGLSLYARRVLIKPDAQELIPNYLRFVI 351
Query 60 GVVDS 64
GVVDS
Sbjct 352 GVVDS 356
Score = 37.0 bits (84), Expect = 0.022, Method: Composition-based stats.
Identities = 25/69 (36%), Positives = 38/69 (55%), Gaps = 6/69 (8%)
Query 65 AAGGSFTVQKDDKYE-----PLGRGTRIILHLK-EDQGEYLEERRLKDLVKKHSEFISFP 118
A G +T D+ YE L GT+I + LK D Y EE R+K+++ K+S F+S P
Sbjct 181 ADGLQWTWNGDNSYEIAETSGLQTGTKIEIRLKVGDSATYAEEDRIKEVINKYSYFVSAP 240
Query 119 IELAVEKTH 127
I + E+ +
Sbjct 241 ILVNGERVN 249
> eco:b0029 ispH, ECK0030, JW0027, lytB, yaaE; 4-hydroxy-3-methylbut-2-enyl
diphosphate reductase, 4Fe-4S protein (EC:1.17.1.2);
K03527 4-hydroxy-3-methylbut-2-enyl diphosphate reductase
[EC:1.17.1.2]
Length=316
Score = 32.7 bits (73), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 32/60 (53%), Gaps = 3/60 (5%)
Query 39 RRAFIMDDCEDIIPEWLNFVK--GVVDSAAGGSFTVQK-DDKYEPLGRGTRIILHLKEDQ 95
+RAF++DD +DI EW+ VK GV A+ VQ + + LG G I L +E+
Sbjct 240 KRAFLIDDAKDIQEEWVKEVKCVGVTAGASAPDILVQNVVARLQQLGGGEAIPLEGREEN 299
> mmu:22378 Wbp2; WW domain binding protein 2
Length=261
Score = 32.3 bits (72), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 30/51 (58%), Gaps = 1/51 (1%)
Query 21 PFDLFETRKKRNNIKLYVRRAFIMDDCEDIIPEW-LNFVKGVVDSAAGGSF 70
P+ + K ++ ++ ++ ++M DCE P + NF+KG+V + AGG +
Sbjct 54 PYRVIFLSKGKDAMQSFMMPFYLMKDCEIKQPVFGANFIKGIVKAEAGGGW 104
Lambda K H
0.318 0.138 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2683748972
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40