bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5495_orf3
Length=128
Score E
Sequences producing significant alignments: (Bits) Value
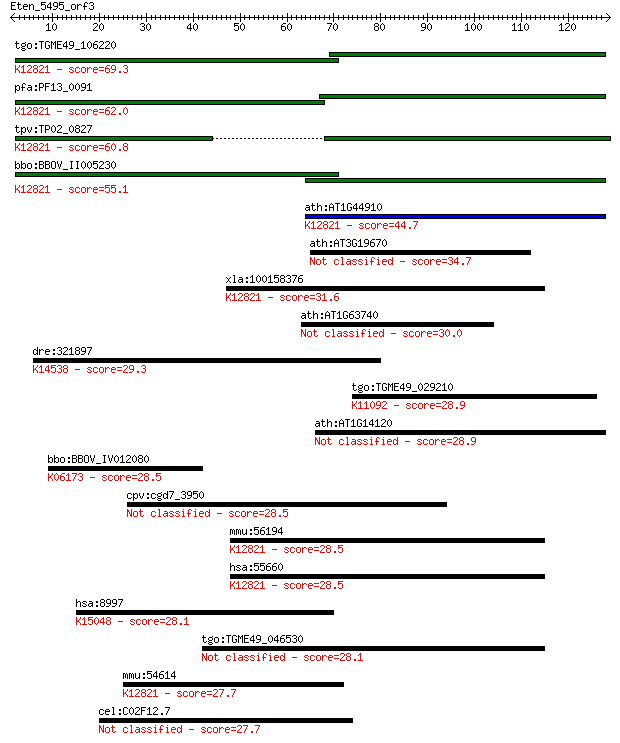
tgo:TGME49_106220 formin binding protein, putative ; K12821 pr... 69.3 3e-12
pfa:PF13_0091 conserved Plasmodium protein, unknown function; ... 62.0 4e-10
tpv:TP02_0827 hypothetical protein; K12821 pre-mRNA-processing... 60.8 1e-09
bbo:BBOV_II005230 18.m06432; WW domain containing protein; K12... 55.1 6e-08
ath:AT1G44910 protein binding; K12821 pre-mRNA-processing fact... 44.7 7e-05
ath:AT3G19670 protein binding 34.7 0.069
xla:100158376 prpf40a, fbp-11, fbp11, flaf1, fnbp3, hip10, hyp... 31.6 0.72
ath:AT1G63740 disease resistance protein (TIR-NBS-LRR class), ... 30.0 1.7
dre:321897 gnl3, MGC123093, id:ibd2914, nstm, wu:fb38a04, wu:f... 29.3 2.8
tgo:TGME49_029210 U2 small nuclear ribonucleoprotein, putative... 28.9 3.7
ath:AT1G14120 2-oxoglutarate-dependent dioxygenase, putative 28.9 3.8
bbo:BBOV_IV012080 23.m06054; tRNA pseudouridine synthase (EC:4... 28.5 5.0
cpv:cgd7_3950 2*FF domain protein (phosphopeptide binding) 28.5 5.2
mmu:56194 Prpf40a, 2810012K09Rik, FBP11, Fnbp11, Fnbp3; PRP40 ... 28.5 5.6
hsa:55660 PRPF40A, FBP-11, FBP11, FLAF1, FLJ20585, FNBP3, HIP-... 28.5 5.9
hsa:8997 KALRN, ARHGEF24, DUET, DUO, FLJ12332, FLJ16443, FLJ18... 28.1 7.2
tgo:TGME49_046530 phosphatidylglycerophosphate synthase, putat... 28.1 7.5
mmu:54614 Prpf40b, 2610317D23Rik, Hypc, MGC91243; PRP40 pre-mR... 27.7 8.4
cel:C02F12.7 tag-278; Temporarily Assigned Gene name family me... 27.7 8.8
> tgo:TGME49_106220 formin binding protein, putative ; K12821
pre-mRNA-processing factor 40
Length=601
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 32/59 (54%), Positives = 45/59 (76%), Gaps = 0/59 (0%)
Query 69 LESDRRWSCFSILTRGERKQTFAEFMGSRGKRAAEEERQKRKKAKDLMVQNLLAWKELS 127
LE+D+RW F ILTRGERKQ+F+EFM R K+ A+ R+KR++A+D + Q L W+EL+
Sbjct 211 LETDKRWESFKILTRGERKQSFSEFMSQRQKKTADSTRKKRQEARDALAQALQNWEELA 269
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 30/69 (43%), Positives = 52/69 (75%), Gaps = 0/69 (0%)
Query 2 LLAWKELSFKTTYIDVADRFHFEEWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRRED 61
L W+EL+ TTYI +AD+ H +EWWT++ E ERD+FFQ++M + ++ ++ K++R++D
Sbjct 262 LQNWEELAPGTTYIAMADKMHEQEWWTFLTEQERDDFFQDYMEEFDKRHRELFKKKRKKD 321
Query 62 VAMLEEILE 70
V +E+IL+
Sbjct 322 VETVEKILD 330
> pfa:PF13_0091 conserved Plasmodium protein, unknown function;
K12821 pre-mRNA-processing factor 40
Length=906
Score = 62.0 bits (149), Expect = 4e-10, Method: Composition-based stats.
Identities = 29/61 (47%), Positives = 44/61 (72%), Gaps = 0/61 (0%)
Query 67 EILESDRRWSCFSILTRGERKQTFAEFMGSRGKRAAEEERQKRKKAKDLMVQNLLAWKEL 126
+ILE+D RWS ILT+GE+KQ F E++ KR E ER+KR+K+++++ Q LL W +L
Sbjct 464 KILEADNRWSSLVILTKGEKKQLFCEYISHVIKRNNENERRKRQKSREIIFQTLLNWDKL 523
Query 127 S 127
+
Sbjct 524 N 524
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 25/66 (37%), Positives = 46/66 (69%), Gaps = 0/66 (0%)
Query 2 LLAWKELSFKTTYIDVADRFHFEEWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRRED 61
LL W +L+ TTY++ A +F+ +EWW W+ E ERDE FQ++M + FK+ +++R++
Sbjct 517 LLNWDKLNECTTYVEFASQFYKQEWWEWITEKERDEVFQDFMDGYKSKFKETRRKKRKQK 576
Query 62 VAMLEE 67
+ +L++
Sbjct 577 MEILKQ 582
> tpv:TP02_0827 hypothetical protein; K12821 pre-mRNA-processing
factor 40
Length=390
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 26/61 (42%), Positives = 45/61 (73%), Gaps = 0/61 (0%)
Query 68 ILESDRRWSCFSILTRGERKQTFAEFMGSRGKRAAEEERQKRKKAKDLMVQNLLAWKELS 127
+LE+D +W FSIL++G++KQ F+EF +R EE+R+K+ +++M++ LL W+ELS
Sbjct 119 LLEADPKWPVFSILSKGDKKQLFSEFCSQIHRRKQEEQRKKKGMLEEVMIRELLNWEELS 178
Query 128 F 128
+
Sbjct 179 Y 179
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 22/42 (52%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 2 LLAWKELSFKTTYIDVADRFHFEEWWTWMQESERDEFFQEWM 43
LL W+ELS+ T Y D + +FH EWW W E RD FQE+M
Sbjct 171 LLNWEELSYATVYADFSKQFHTAEWWDWGDEVTRDAIFQEFM 212
> bbo:BBOV_II005230 18.m06432; WW domain containing protein; K12821
pre-mRNA-processing factor 40
Length=457
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 28/64 (43%), Positives = 40/64 (62%), Gaps = 0/64 (0%)
Query 64 MLEEILESDRRWSCFSILTRGERKQTFAEFMGSRGKRAAEEERQKRKKAKDLMVQNLLAW 123
M +LE + RW F+ILT+GE+KQ F+EF +R EE R+KR DL++ L W
Sbjct 117 MAVRLLEVNERWPKFAILTKGEKKQLFSEFTSQAQRRHHEEMRRKRGMIGDLIINELEKW 176
Query 124 KELS 127
+EL+
Sbjct 177 EELT 180
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 29/69 (42%), Positives = 41/69 (59%), Gaps = 0/69 (0%)
Query 2 LLAWKELSFKTTYIDVADRFHFEEWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRRED 61
L W+EL+ TTY++ A+R H EWWTW E RD FQE M + K++ ++RRR
Sbjct 173 LEKWEELTPYTTYVEFAERCHTREWWTWADEKTRDGIFQETMERMDHELKERQRERRRVS 232
Query 62 VAMLEEILE 70
+ LE +E
Sbjct 233 MEKLEAEME 241
> ath:AT1G44910 protein binding; K12821 pre-mRNA-processing factor
40
Length=926
Score = 44.7 bits (104), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 28/64 (43%), Positives = 41/64 (64%), Gaps = 3/64 (4%)
Query 64 MLEEILESDRRWSCFSILTRGERKQTFAEFMGSRGKRAAEEERQKRKKAKDLMVQNLLAW 123
L+EI+ D+R+ L GERKQ F E++G R K AEE R+++KKA++ V+ L
Sbjct 429 TLKEIVH-DKRYGALRTL--GERKQAFNEYLGQRKKVEAEERRRRQKKAREEFVKMLEEC 485
Query 124 KELS 127
+ELS
Sbjct 486 EELS 489
> ath:AT3G19670 protein binding
Length=992
Score = 34.7 bits (78), Expect = 0.069, Method: Compositional matrix adjust.
Identities = 23/47 (48%), Positives = 31/47 (65%), Gaps = 4/47 (8%)
Query 65 LEEILESDRRWSCFSILTRGERKQTFAEFMGSRGKRAAEEERQKRKK 111
+ EI+ +D+R+ L GERKQ F EF+ + KRAAEEER R+K
Sbjct 476 MREII-NDKRYGALRTL--GERKQAFNEFL-LQTKRAAEEERLARQK 518
> xla:100158376 prpf40a, fbp-11, fbp11, flaf1, fnbp3, hip10, hypa,
prp40; PRP40 pre-mRNA processing factor 40 homolog A; K12821
pre-mRNA-processing factor 40
Length=487
Score = 31.6 bits (70), Expect = 0.72, Method: Composition-based stats.
Identities = 22/71 (30%), Positives = 39/71 (54%), Gaps = 5/71 (7%)
Query 47 EQMFKDKIKQRRREDVAMLEEILE---SDRRWSCFSILTRGERKQTFAEFMGSRGKRAAE 103
+Q FK+ +K++R A E+ ++ +D R+S + L+ E+KQ + + K E
Sbjct 334 KQAFKELLKEKRVPSNATWEQAMKMIINDPRYSALAKLS--EKKQAYNAYKVQTEKEEKE 391
Query 104 EERQKRKKAKD 114
E R K K+AK+
Sbjct 392 EARLKYKEAKE 402
> ath:AT1G63740 disease resistance protein (TIR-NBS-LRR class),
putative
Length=992
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 19/48 (39%), Positives = 25/48 (52%), Gaps = 7/48 (14%)
Query 63 AMLEEILESDRRWSCF-------SILTRGERKQTFAEFMGSRGKRAAE 103
AMLEE+LES R WSC S++T T E MG+ +R +
Sbjct 688 AMLEEMLESIRLWSCLETLVVYGSVITHNFWAVTLIEKMGTDIERIPD 735
> dre:321897 gnl3, MGC123093, id:ibd2914, nstm, wu:fb38a04, wu:fc55d07,
zgc:123093; guanine nucleotide binding protein-like
3 (nucleolar); K14538 nuclear GTP-binding protein
Length=561
Score = 29.3 bits (64), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 20/74 (27%), Positives = 36/74 (48%), Gaps = 6/74 (8%)
Query 6 KELSFKTTYIDVADRFHFEEWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRREDVAML 65
K+L F ID+ + + E+W ++ E+E F +F + KD+ Q++R+
Sbjct 174 KKLLFILNKIDLVPKDNLEKWLHFL-EAECPTF----LFKSSMQLKDRTVQQKRQQRGT- 227
Query 66 EEILESDRRWSCFS 79
+L+ R SCF
Sbjct 228 NAVLDHSRAASCFG 241
> tgo:TGME49_029210 U2 small nuclear ribonucleoprotein, putative
(EC:3.1.3.16); K11092 U2 small nuclear ribonucleoprotein
A'
Length=254
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 28/52 (53%), Gaps = 0/52 (0%)
Query 74 RWSCFSILTRGERKQTFAEFMGSRGKRAAEEERQKRKKAKDLMVQNLLAWKE 125
R+ +S +T+ ER+Q + F G +G + A E RK L V ++ A +E
Sbjct 142 RFLDYSRVTQQEREQAHSTFKGEKGAKLAHEIAPPRKSHAALGVSSVGAAEE 193
> ath:AT1G14120 2-oxoglutarate-dependent dioxygenase, putative
Length=312
Score = 28.9 bits (63), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 30/62 (48%), Gaps = 3/62 (4%)
Query 66 EEILESDRRWSCFSILTRGERKQTFAEFMGSRGKRAAEEERQKRKKAKDLMVQNLLAWKE 125
E+I E+ RW CF+++ G AE M + E + + + D+++ N +K
Sbjct 23 EKIREASERWGCFTVINHGVSLSLMAE-MKKTVRDLHERPYEMKLRNTDVLLGN--GYKP 79
Query 126 LS 127
LS
Sbjct 80 LS 81
> bbo:BBOV_IV012080 23.m06054; tRNA pseudouridine synthase (EC:4.2.1.70);
K06173 tRNA pseudouridine synthase A [EC:5.4.99.12]
Length=414
Score = 28.5 bits (62), Expect = 5.0, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 21/35 (60%), Gaps = 2/35 (5%)
Query 9 SFKTT--YIDVADRFHFEEWWTWMQESERDEFFQE 41
+FK T Y ++A F + W TWM++ R+ F+ E
Sbjct 358 NFKRTRIYPEIASTFETDVWKTWMEKLLRNPFYLE 392
> cpv:cgd7_3950 2*FF domain protein (phosphopeptide binding)
Length=431
Score = 28.5 bits (62), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 39/80 (48%), Gaps = 14/80 (17%)
Query 26 WWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRREDV----------AMLEEILESDRRW 75
WW +++ E+DE FQE + ++E+ F ++ E+V + LES++
Sbjct 113 WWNNIEDKEKDEIFQELVEEHEKNFIRTLEPNYEENVNDFFELLRLETTIFPFLESNKAE 172
Query 76 SCFSILTRGER--KQTFAEF 93
S SI+ GE K+ F F
Sbjct 173 S--SIIRNGESIEKKNFTGF 190
> mmu:56194 Prpf40a, 2810012K09Rik, FBP11, Fnbp11, Fnbp3; PRP40
pre-mRNA processing factor 40 homolog A (yeast); K12821 pre-mRNA-processing
factor 40
Length=953
Score = 28.5 bits (62), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 38/70 (54%), Gaps = 5/70 (7%)
Query 48 QMFKDKIKQRRREDVAMLEEILE---SDRRWSCFSILTRGERKQTFAEFMGSRGKRAAEE 104
Q FK+ +K++R A E+ ++ +D R+S + L+ E+KQ F + K EE
Sbjct 394 QAFKELLKEKRVPSNASWEQAMKMIINDPRYSALAKLS--EKKQAFNAYKVQTEKEEKEE 451
Query 105 ERQKRKKAKD 114
R K K+AK+
Sbjct 452 ARSKYKEAKE 461
> hsa:55660 PRPF40A, FBP-11, FBP11, FLAF1, FLJ20585, FNBP3, HIP-10,
HIP10, HYPA, NY-REN-6, Prp40; PRP40 pre-mRNA processing
factor 40 homolog A (S. cerevisiae); K12821 pre-mRNA-processing
factor 40
Length=930
Score = 28.5 bits (62), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 38/70 (54%), Gaps = 5/70 (7%)
Query 48 QMFKDKIKQRRREDVAMLEEILE---SDRRWSCFSILTRGERKQTFAEFMGSRGKRAAEE 104
Q FK+ +K++R A E+ ++ +D R+S + L+ E+KQ F + K EE
Sbjct 371 QAFKELLKEKRVPSNASWEQAMKMIINDPRYSALAKLS--EKKQAFNAYKVQTEKEEKEE 428
Query 105 ERQKRKKAKD 114
R K K+AK+
Sbjct 429 ARSKYKEAKE 438
> hsa:8997 KALRN, ARHGEF24, DUET, DUO, FLJ12332, FLJ16443, FLJ18196,
FLJ18623, HAPIP, TRAD; kalirin, RhoGEF kinase (EC:2.7.11.1);
K15048 kalirin [EC:2.7.11.1]
Length=2986
Score = 28.1 bits (61), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 15/55 (27%), Positives = 31/55 (56%), Gaps = 1/55 (1%)
Query 15 IDVADRFHFEEWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRREDVAMLEEIL 69
+ V+ H +E WTWM++ ++ E ++ D+ ++ IKQ +++ A L+ L
Sbjct 642 MSVSFHTHTKELWTWMEDLQK-EMLEDVCADSVDAVQELIKQFQQQQTATLDATL 695
> tgo:TGME49_046530 phosphatidylglycerophosphate synthase, putative
(EC:2.7.8.8 2.7.8.5)
Length=986
Score = 28.1 bits (61), Expect = 7.5, Method: Composition-based stats.
Identities = 24/92 (26%), Positives = 38/92 (41%), Gaps = 23/92 (25%)
Query 42 WMFDNEQ--MFKDKIKQRRREDVAMLEEI-----------------LESDRRWSCFSILT 82
W F + +F D+ Q RR+ V L +I ++ R CFS +
Sbjct 778 WTFHTKGIWLFADEKTQERRKAVGFLPDIPDRTACGATETARHRGLVDPQREEGCFSFSS 837
Query 83 RGERKQTFAEFMGSRGKRAAEEERQKRKKAKD 114
G R + +EF+ R + RQ+ KAK+
Sbjct 838 AGSRGRNSSEFL----FRTSPSVRQRTCKAKE 865
> mmu:54614 Prpf40b, 2610317D23Rik, Hypc, MGC91243; PRP40 pre-mRNA
processing factor 40 homolog B (yeast); K12821 pre-mRNA-processing
factor 40
Length=873
Score = 27.7 bits (60), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 25 EWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRREDVAMLEEILES 71
E W + E ER E + + +F + K++ KQ RR ++ L+ IL+
Sbjct 377 EVWAVVPERERKEVYDDVLFFLAKKEKEQAKQLRRRNIQALKSILDG 423
> cel:C02F12.7 tag-278; Temporarily Assigned Gene name family
member (tag-278)
Length=1130
Score = 27.7 bits (60), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 32/58 (55%), Gaps = 6/58 (10%)
Query 20 RFHFEEWWTWMQESERDE----FFQEWMFDNEQMFKDKIKQRRREDVAMLEEILESDR 73
+ H E + Q +R+E FQE MF+ EQ+ +K +Q E A +E++ ++D+
Sbjct 900 KLHEEMYMQKTQNEKRNEEQSKLFQELMFEKEQLEAEKAEQSHIE--AEVEQVFQADK 955
Lambda K H
0.322 0.132 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2049573556
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40