bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
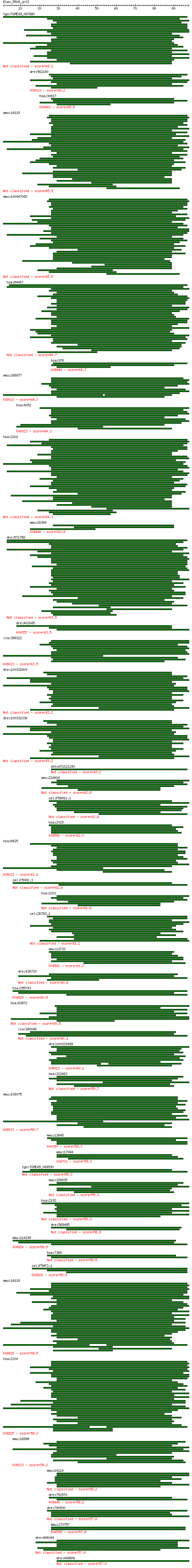
Query= Eten_5508_orf2
Length=97
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_067680 microneme protein, putative (EC:1.14.99.1 3.... 68.2 6e-12
dre:562180 ltbp1; latent transforming growth factor beta bindi... 66.2 2e-11
hsa:30817 EMR2, CD312, DKFZp781B135; egf-like module containin... 65.5 4e-11
mmu:14119 Fbn2, BC063774, Fib-2, KIAA4226, mKIAA4226, sne, sy;... 65.5 4e-11
mmu:100047082 fibrillin-2-like 65.5 4e-11
hsa:84467 FBN3, KIAA1776; fibrillin 3 64.7
hsa:976 CD97, TM7LN1; CD97 molecule; K08446 CD97 molecule 64.7 7e-11
mmu:268977 Ltbp1, 9430031G15Rik, 9830146M04, Ltbp-1, Tgfb; lat... 64.7 7e-11
hsa:4052 LTBP1, MGC163161; latent transforming growth factor b... 64.3 8e-11
hsa:2201 FBN2, CCA, DA9; fibrillin 2 64.3
mmu:26364 Cd97, AA409984, TM7LN1; CD97 antigen; K08446 CD97 mo... 63.9 1e-10
dre:571786 fbn2b, fbn2; fibrillin 2b 63.5 1e-10
dre:403045 egf; epidermal growth factor; K04357 epidermal grow... 63.5 1e-10
xla:398322 ltbp1, ltgf-1; latent transforming growth factor be... 63.5 2e-10
dre:100332400 novel EGF domain containing protein-like 63.2 2e-10
dre:100332336 novel EGF domain containing protein-like 63.2 2e-10
ath:AT1G21240 WAK3; WAK3 (wall associated kinase 3); kinase/ p... 63.2 2e-10
mmu:216616 Efemp1, MGC37612; epidermal growth factor-containin... 62.8 3e-10
cel:F56H11.1 hypothetical protein 62.8 3e-10
hsa:2015 EMR1, TM7LN3; egf-like module containing, mucin-like,... 62.0 4e-10
hsa:8425 LTBP4, FLJ46318, FLJ90018, LTBP-4, LTBP4L, LTBP4S; la... 61.6 5e-10
cel:C50H2.3 mec-9; MEChanosensory abnormality family member (m... 61.6 6e-10
hsa:2202 EFEMP1, DHRD, DRAD, FBLN3, FBNL, FLJ35535, MGC111353,... 61.6 6e-10
cel:ZK783.1 fbn-1; FiBrilliN homolog family member (fbn-1) 61.2 7e-10
mmu:13733 Emr1, DD7A5-7, EGF-TM7, F4/80, Gpf480, Ly71, TM7LN3;... 61.2 7e-10
dre:436730 egfl6, sb:cb3, wu:fc66b04, zgc:92328; EGF-like-doma... 60.8 1e-09
hsa:255743 NPNT, EGFL6L, POEM; nephronectin; K06824 nephronectin 60.5 1e-09
hsa:83872 HMCN1, ARMD1, FBLN6, FIBL-6, FIBL6; hemicentin 1 60.5
xla:380046 egfl6, MGC53438; EGF-like-domain, multiple 6 60.1
dre:100333499 latent transforming growth factor beta binding p... 60.1 2e-09
hsa:222663 SCUBE3, CEGF3, DKFZp686B09105, DKFZp686B1223, DKFZp... 59.7 2e-09
mmu:108075 Ltbp4, 2310046A13Rik, MGC175966; latent transformin... 59.7 2e-09
mmu:13645 Egf, AI790464; epidermal growth factor; K04357 epide... 59.3 3e-09
mmu:17064 Cd93, 6030404G09Rik, AA145088, AA4.1, AW555904, C1qr... 59.3 3e-09
tgo:TGME49_069930 hypothetical protein 59.3 3e-09
mmu:268935 Scube3, CEGF3, D030038I21Rik; signal peptide, CUB d... 59.3 3e-09
hsa:2192 FBLN1, FBLN, FIBL1; fibulin 1 59.3
dre:569485 si:ch211-29n12.1 58.9 4e-09
mmu:114249 Npnt, 1110009H02Rik, AA682063, AI314031, Nctn, POEM... 58.5 4e-09
hsa:7369 UMOD, ADMCKD2, FJHN, HNFJ, HNFJ1, MCKD2, THGP, THP; u... 58.5 5e-09
cel:F54F3.1 nid-1; NIDogen (basement membrane protein family m... 58.5 5e-09
mmu:14118 Fbn1, AI536462, B430209H23, Fib-1, Tsk; fibrillin 1;... 58.5 6e-09
hsa:2200 FBN1, FBN, MASS, MFS1, OCTD, SGS, SSKS, WMS; fibrilli... 58.2 6e-09
mmu:16998 Ltbp3, Ltbp2, mFLJ00070; latent transforming growth ... 58.2 6e-09
mmu:14114 Fbln1; fibulin 1 58.2
dre:792970 CD97 antigen-like; K08446 CD97 molecule 58.2 7e-09
dre:790930 nell2; zgc:158375 57.8 8e-09
mmu:170757 Eltd1, 1110033N21Rik, ETL1, Etl; EGF, latrophilin s... 57.8 9e-09
dre:494044 efemp2, wu:fb22f10, wu:fc49d09, zgc:103659; EGF-con... 57.4 1e-08
dre:449806 fbln5, zgc:103575; fibulin 5 57.0
> tgo:TGME49_067680 microneme protein, putative (EC:1.14.99.1
3.4.21.42 2.7.10.1 1.11.1.8 3.4.21.6 3.4.21.21)
Length=2182
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 38/96 (39%), Positives = 50/96 (52%), Gaps = 1/96 (1%)
Query 1 LPMVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDEC 60
L V CV + + + +A+C NTEGS+ C CN GY C ++DEC
Sbjct 427 LGKVSSPCVDIDECDREHPTHDCDSNATCTNTEGSFTCACNTGYTGEGRGADTCTEIDEC 486
Query 61 AAGTAECHVSAQCVNVDGSFECTCMAGFEAADAKTC 96
A GTA C A C N GSF+CTC+ G+ + D TC
Sbjct 487 ADGTANCAAEATCTNTPGSFKCTCLEGY-SGDGFTC 521
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 34/69 (49%), Positives = 41/69 (59%), Gaps = 0/69 (0%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
S H+ C N GSY C C+ GYE H C+D+DECAAGTA ++ CVN GS+E
Sbjct 1015 SEHSQCRNLPGSYECVCDAGYEKVEGSEHLCQDIDECAAGTATIPNNSNCVNTAGSYEFA 1074
Query 84 CMAGFEAAD 92
C GFE D
Sbjct 1075 CKPGFEHKD 1083
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 33/65 (50%), Positives = 40/65 (61%), Gaps = 1/65 (1%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT GSY C+C PGY P S DG C D+DECA C +QC N+ GS+EC C AG+
Sbjct 977 CTNTMGSYTCSCLPGYTP-SDDGRVCTDIDECATENGGCSEHSQCRNLPGSYECVCDAGY 1035
Query 89 EAADA 93
E +
Sbjct 1036 EKVEG 1040
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 34/71 (47%), Positives = 42/71 (59%), Gaps = 3/71 (4%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
A+C NT GSY C+CNPG+ S DGH C D++EC +C C N GSF C C
Sbjct 750 ATCHNTAGSYTCSCNPGF---SGDGHECADINECETNAHDCGSHTTCENTVGSFVCNCKE 806
Query 87 GFEAADAKTCK 97
GF +D KTC+
Sbjct 807 GFVHSDEKTCR 817
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 36/71 (50%), Positives = 44/71 (61%), Gaps = 3/71 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H +C NT GS+VC C G+ SD C+DVDECA +C V A C N +GSFEC+C
Sbjct 790 HTTCENTVGSFVCNCKEGF--VHSDEKTCRDVDECAENKHDCSVHATCNNTEGSFECSCK 847
Query 86 AGFEAADAKTC 96
AGFE + K C
Sbjct 848 AGFE-GNGKEC 857
Score = 61.6 bits (148), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 32/71 (45%), Positives = 44/71 (61%), Gaps = 4/71 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
HA+C NT+GSY C C G+ + DG C+D++EC G EC +A C N+ GS+ C C
Sbjct 1441 HATCENTDGSYHCACGSGF---TGDGFTCEDINECETGEHECDSNATCENIVGSYSCHCP 1497
Query 86 AGFEAADAKTC 96
GF A D ++C
Sbjct 1498 TGF-AGDGRSC 1507
Score = 61.2 bits (147), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 37/70 (52%), Positives = 39/70 (55%), Gaps = 4/70 (5%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
A+C NT GS+ CTC GY S DG C D DEC A CH SA C N GSF C C A
Sbjct 497 ATCTNTPGSFKCTCLEGY---SGDGFTCSDNDECQQEPAPCHQSATCQNTPGSFTCACNA 553
Query 87 GFEAADAKTC 96
GF D TC
Sbjct 554 GFR-GDGHTC 562
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 43/98 (43%), Positives = 52/98 (53%), Gaps = 15/98 (15%)
Query 12 PQGEASAAHSSTSLHAS-------CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGT 64
P+G A T H S C NTEGSY C+C GY + DG C+D+DECA+G
Sbjct 687 PEGCADRDECQTENHCSTDENGGICTNTEGSYTCSCKEGYRQLA-DG-TCEDIDECASG- 743
Query 65 AECHVSAQCVNVDGSFECTCMAGF-----EAADAKTCK 97
ECH SA C N GS+ C+C GF E AD C+
Sbjct 744 HECHESATCHNTAGSYTCSCNPGFSGDGHECADINECE 781
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 35/74 (47%), Positives = 41/74 (55%), Gaps = 5/74 (6%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
S HA C + + VCTC PGYE DG C D+DECA T C A C N DGS+ C
Sbjct 1399 SPHADCEHLD-KIVCTCRPGYE---GDGITCTDIDECALNTDNCDSHATCENTDGSYHCA 1454
Query 84 CMAGFEAADAKTCK 97
C +GF D TC+
Sbjct 1455 CGSGF-TGDGFTCE 1467
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 32/63 (50%), Positives = 41/63 (65%), Gaps = 3/63 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H++C NTEGSY C C GY A S+ +C+DV+ECA + C ++ CVN GSFEC C
Sbjct 279 HSTCRNTEGSYDCDCKTGY--AMSETGSCEDVNECATENS-CPENSSCVNTAGSFECVCN 335
Query 86 AGF 88
GF
Sbjct 336 EGF 338
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 34/80 (42%), Positives = 40/80 (50%), Gaps = 7/80 (8%)
Query 13 QGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQ 72
Q E + H S A+C NT GS+ C CN G+ DGH C D+DECA C A
Sbjct 528 QQEPAPCHQS----ATCQNTPGSFTCACNAGF---RGDGHTCGDIDECAEDPNACGAHAV 580
Query 73 CVNVDGSFECTCMAGFEAAD 92
C N GSF C C G+ D
Sbjct 581 CRNTVGSFSCNCEEGYGNLD 600
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 30/66 (45%), Positives = 37/66 (56%), Gaps = 4/66 (6%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT GSY C+C PGY+ GH C D++EC C +A C N GS+ C C AGF
Sbjct 1196 CTNTPGSYTCSCKPGYD---QQGHDCVDINECTT-QEPCGDNADCENTSGSYICKCKAGF 1251
Query 89 EAADAK 94
E D +
Sbjct 1252 EMRDNQ 1257
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 32/65 (49%), Positives = 39/65 (60%), Gaps = 3/65 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGH-ACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
HA C NT GS+ C C GY + D H AC D++EC A +A CVN DGSFE +C
Sbjct 578 HAVCRNTVGSFSCNCEEGY--GNLDEHRACHDINECEAEPERIPPNATCVNTDGSFEWSC 635
Query 85 MAGFE 89
AG+E
Sbjct 636 NAGYE 640
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 29/63 (46%), Positives = 38/63 (60%), Gaps = 1/63 (1%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
++ C +TEGSY C CN GY+ + +C D DEC A C +A C N+ GSF CTC
Sbjct 1316 NSHCVDTEGSYKCDCNSGYKQDPENPDSCIDRDECEIEGA-CDENADCTNLPGSFSCTCR 1374
Query 86 AGF 88
AG+
Sbjct 1375 AGY 1377
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 47/89 (52%), Gaps = 4/89 (4%)
Query 1 LPMVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDEC 60
M QCV + + A+ + + C NT GSY C C G+ DG C + +EC
Sbjct 1251 FEMRDNQCVDIDEC-ATNTNECHNHRGRCINTHGSYTCECIAGF---IGDGKICINKNEC 1306
Query 61 AAGTAECHVSAQCVNVDGSFECTCMAGFE 89
+G EC ++ CV+ +GS++C C +G++
Sbjct 1307 QSGDFECGPNSHCVDTEGSYKCDCNSGYK 1335
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 42/70 (60%), Gaps = 5/70 (7%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDG--SFE 81
S+HA+C NTEGS+ C+C G+E +G C D+ C+AG ++C +A C + +
Sbjct 830 SVHATCNNTEGSFECSCKAGFE---GNGKECSDIQFCSAGRSDCAANADCAENEAGTDYA 886
Query 82 CTCMAGFEAA 91
C+C AG+ +
Sbjct 887 CSCHAGYRGS 896
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 2/77 (2%)
Query 18 AAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVD 77
A +S ++SC NT GS+ C CN G+ +++ C+D+DECA C +A C N
Sbjct 312 ATENSCPENSSCVNTAGSFECVCNEGFR-MNAETQQCEDIDECAE-EGGCSANATCTNSV 369
Query 78 GSFECTCMAGFEAADAK 94
GS+ C+C G++ +
Sbjct 370 GSYSCSCPEGYKGEGTR 386
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 28/75 (37%), Positives = 36/75 (48%), Gaps = 4/75 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAA--GTAECHVSAQCVNVDGSFECT 83
HA+C N + C C+ GY C D+DEC T +C +A C N +GSF C
Sbjct 406 HATCRNEAVGFTCICDAGYTGLGKVSSPCVDIDECDREHPTHDCDSNATCTNTEGSFTCA 465
Query 84 CMAGF--EAADAKTC 96
C G+ E A TC
Sbjct 466 CNTGYTGEGRGADTC 480
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 28/74 (37%), Positives = 39/74 (52%), Gaps = 3/74 (4%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E A +A+C NT+GS+ +CN GYE G C+ ++ CA G H S Q V
Sbjct 610 ECEAEPERIPPNATCVNTDGSFEWSCNAGYEHV---GSQCQKINFCARGFCSPHASCQEV 666
Query 75 NVDGSFECTCMAGF 88
+ S+ECTC G+
Sbjct 667 SNGTSYECTCQPGY 680
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 33/65 (50%), Gaps = 0/65 (0%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
S +A+C N+ GSY C+C GY+ + C +D C G +C A C N F C
Sbjct 360 SANATCTNSVGSYSCSCPEGYKGEGTRDSPCNKIDYCGEGLHDCGEHATCRNEAVGFTCI 419
Query 84 CMAGF 88
C AG+
Sbjct 420 CDAGY 424
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 37/77 (48%), Gaps = 3/77 (3%)
Query 24 SLHASCANTEGSYVCTCNPGY---EPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSF 80
S HA C C C GY E A + C D DECA T +C + C N +GS+
Sbjct 230 SEHAVCFADGLQAKCRCEKGYDGKEGAGTQDDPCIDRDECATNTHQCPAHSTCRNTEGSY 289
Query 81 ECTCMAGFEAADAKTCK 97
+C C G+ ++ +C+
Sbjct 290 DCDCKTGYAMSETGSCE 306
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 40/79 (50%), Gaps = 7/79 (8%)
Query 17 SAAHSSTSLHASCANTEG--SYVCTCNPGYEPA--SSDGHA--CKDVDECAAGTAEC-HV 69
SA S + +A CA E Y C+C+ GY + +S G A C D+DEC G C
Sbjct 864 SAGRSDCAANADCAENEAGTDYACSCHAGYRGSGHTSKGAADGCVDIDECTEGVDTCPRQ 923
Query 70 SAQCVNVDGSFECTCMAGF 88
+CVN GS+ C C G+
Sbjct 924 GGRCVNTPGSYRCECEEGY 942
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 30/81 (37%), Positives = 35/81 (43%), Gaps = 10/81 (12%)
Query 24 SLHASCANTEG--SYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQ-----CVNV 76
S HASC SY CTC PGY C D DEC E H S C N
Sbjct 658 SPHASCQEVSNGTSYECTCQPGYVGDGVGPEGCADRDECQ---TENHCSTDENGGICTNT 714
Query 77 DGSFECTCMAGFEAADAKTCK 97
+GS+ C+C G+ TC+
Sbjct 715 EGSYTCSCKEGYRQLADGTCE 735
Score = 35.8 bits (81), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 33/63 (52%), Gaps = 5/63 (7%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAEC--HVSAQCVNVDGSFECTCMA 86
C NT GS+ C+C GY + C+++DECA ++ C N GS+ C+C
Sbjct 1153 CENTPGSFNCSCANGY---LLNNGVCEEIDECAGSSSNTCADEGGICTNTPGSYTCSCKP 1209
Query 87 GFE 89
G++
Sbjct 1210 GYD 1212
Score = 34.3 bits (77), Expect = 0.099, Method: Compositional matrix adjust.
Identities = 27/76 (35%), Positives = 39/76 (51%), Gaps = 7/76 (9%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E +A ++ +++C NT GSY C PG+E + AC +D C G C+ A C
Sbjct 1050 ECAAGTATIPNNSNCVNTAGSYEFACKPGFEHKDN---ACSKIDYCGRGG--CNSLATCE 1104
Query 75 NV-DGS-FECTCMAGF 88
DG+ + CTC GF
Sbjct 1105 ETADGTDYVCTCPKGF 1120
Score = 33.9 bits (76), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 25/59 (42%), Positives = 30/59 (50%), Gaps = 8/59 (13%)
Query 36 YVCTCNPGY----EPASSDGHACKDVDECAAGTAECHVSAQ--CVNVDGSFECTCMAGF 88
YVCTC G+ E +DG C DVDECA + S C N GSF C+C G+
Sbjct 1112 YVCTCPKGFVTQNEGRGADG--CTDVDECAENGCAAYGSEGVICENTPGSFNCSCANGY 1168
> dre:562180 ltbp1; latent transforming growth factor beta binding
protein 1; K08023 latent transforming growth factor beta
binding protein
Length=1154
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 43/72 (59%), Gaps = 1/72 (1%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
+ C NT+GS++C C PG++ SS G C+DVDEC C QCVN+ GS++C C
Sbjct 867 IQGHCQNTQGSFICNCEPGFK-LSSSGDQCEDVDECLVIPQTCDGVGQCVNILGSYQCNC 925
Query 85 MAGFEAADAKTC 96
G+ ++ +C
Sbjct 926 PQGYRQVNSTSC 937
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 28/68 (41%), Positives = 39/68 (57%), Gaps = 3/68 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H C+N EG +VCTC+ G+ P + DG +C D+DEC C + C N GSF C C
Sbjct 827 HGQCSNLEGYFVCTCDEGFLP-TPDGKSCTDIDECQDDNV-C-IQGHCQNTQGSFICNCE 883
Query 86 AGFEAADA 93
GF+ + +
Sbjct 884 PGFKLSSS 891
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 34/62 (54%), Gaps = 2/62 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C N GSY C C GY +S +C DVDEC T C + +C+N +GSF C C A
Sbjct 912 GQCVNILGSYQCNCPQGYRQVNST--SCLDVDECEDDTQLCIPNGECLNTEGSFLCVCDA 969
Query 87 GF 88
GF
Sbjct 970 GF 971
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 30/83 (36%), Positives = 42/83 (50%), Gaps = 2/83 (2%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E S ++ H +C NT GS++C C+PG++ S DG C DV+EC C A C
Sbjct 983 ECSLNENTCGSHGTCQNTIGSFICQCDPGFK-DSQDGQGCVDVNECELLGNVCG-EATCE 1040
Query 75 NVDGSFECTCMAGFEAADAKTCK 97
N +G+F C C + D K
Sbjct 1041 NREGTFLCLCPDDLQEFDPMLGK 1063
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 38/72 (52%), Gaps = 4/72 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NTEGS++C C+ G+ S C+DVDEC+ C C N GSF C C
Sbjct 953 NGECLNTEGSFLCVCDAGF---VSTRTGCQDVDECSLNENTCGSHGTCQNTIGSFICQCD 1009
Query 86 AGF-EAADAKTC 96
GF ++ D + C
Sbjct 1010 PGFKDSQDGQGC 1021
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/60 (45%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT SY C C+ GY S+G C DVDECA + C + +C N G F C C GF
Sbjct 707 CYNTAESYTCFCDDGYR-TDSEGTTCVDVDECAENPSLC-ANGRCQNTPGGFLCVCELGF 764
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/78 (37%), Positives = 40/78 (51%), Gaps = 4/78 (5%)
Query 18 AAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVD 77
A + S + C NT G ++C C G+ P + G +C D+DEC A C S C N
Sbjct 738 AENPSLCANGRCQNTPGGFLCVCELGFMP-NERGSSCSDIDECQNRRA-C-PSGLCYNTL 794
Query 78 GSFECT-CMAGFEAADAK 94
GSF C+ C GFE + +
Sbjct 795 GSFMCSPCPDGFEGKNGQ 812
Score = 32.0 bits (71), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 13/24 (54%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 26 HASCANTEGSYVCTCNPGYEPASS 49
+ +C NT GSY C CNPG+ P S
Sbjct 505 NGNCINTVGSYRCICNPGFVPDPS 528
> hsa:30817 EMR2, CD312, DKFZp781B135; egf-like module containing,
mucin-like, hormone receptor-like 2; K08443 egf-like module
containing, mucin-like, hormone receptor-like 2
Length=823
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 38/64 (59%), Gaps = 0/64 (0%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ +C NT GSY C C PG++ D C DV+EC +G CH S C+N GS++C C
Sbjct 133 YGTCVNTLGSYTCQCLPGFKLKPEDPKLCTDVNECTSGQNPCHSSTHCLNNVGSYQCRCR 192
Query 86 AGFE 89
G++
Sbjct 193 PGWQ 196
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 34/79 (43%), Positives = 41/79 (51%), Gaps = 9/79 (11%)
Query 27 ASCANTEGSYVCTCNPGYEPAS-------SDGHACKDVDECAAGTAECHVSAQCVNVDGS 79
+ C NTEGSY C C+PGYEP S + C+DVDEC C CVN GS
Sbjct 83 SDCWNTEGSYDCVCSPGYEPVSGAKTFKNESENTCQDVDECQQNPRLCKSYGTCVNTLGS 142
Query 80 FECTCMAGFEAA--DAKTC 96
+ C C+ GF+ D K C
Sbjct 143 YTCQCLPGFKLKPEDPKLC 161
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 30/75 (40%), Positives = 40/75 (53%), Gaps = 9/75 (12%)
Query 20 HSSTSLHASCANTEGSYVCTCNPGYEPASS-----DGHACKDVDECAAGTAECHVSAQCV 74
HSST C N GSY C C PG++P + C+DVDEC++G +C S C
Sbjct 175 HSSTH----CLNNVGSYQCRCRPGWQPIPGSPNGPNNTVCEDVDECSSGQHQCDSSTVCF 230
Query 75 NVDGSFECTCMAGFE 89
N GS+ C C G++
Sbjct 231 NTVGSYSCRCRPGWK 245
Score = 31.2 bits (69), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 16/28 (57%), Gaps = 0/28 (0%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKD 56
C NT GSY C C PG++P + KD
Sbjct 229 CFNTVGSYSCRCRPGWKPRHGIPNNQKD 256
> mmu:14119 Fbn2, BC063774, Fib-2, KIAA4226, mKIAA4226, sne, sy;
fibrillin 2
Length=2907
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 34/72 (47%), Positives = 44/72 (61%), Gaps = 4/72 (5%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
+HASC N GS+ C+C G+ +G C D+DECA GT +C ++AQCVN GS+ C C
Sbjct 1373 MHASCLNVPGSFKCSCREGW---VGNGIKCIDLDECANGTHQCSINAQCVNTPGSYRCAC 1429
Query 85 MAGFEAADAKTC 96
GF D TC
Sbjct 1430 SEGF-TGDGFTC 1440
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 34/75 (45%), Positives = 48/75 (64%), Gaps = 5/75 (6%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
S++A C NT GSY C C+ G+ + DG C DVDECA T C + QC+NV G++ C
Sbjct 1413 SINAQCVNTPGSYRCACSEGF---TGDGFTCSDVDECAENTNLCE-NGQCLNVPGAYRCE 1468
Query 84 CMAGF-EAADAKTCK 97
C GF A+D+++C+
Sbjct 1469 CEMGFTPASDSRSCQ 1483
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 41/72 (56%), Gaps = 3/72 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT GSY C C+ G++ + AC D+DEC C + +CVN DGSF+C C
Sbjct 539 NGDCVNTPGSYYCKCHAGFQRTPTK-QACIDIDECIQNGVLCK-NGRCVNTDGSFQCICN 596
Query 86 AGFE-AADAKTC 96
AGFE D K C
Sbjct 597 AGFELTTDGKNC 608
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 41/72 (56%), Gaps = 4/72 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N G+Y C+CNPGY+ A+ D C D+DEC C QC N +GS+EC+C
Sbjct 1206 NGKCVNMIGTYQCSCNPGYQ-ATPDRQGCTDIDECMIMNGGC--DTQCTNSEGSYECSCS 1262
Query 86 AGFE-AADAKTC 96
G+ D ++C
Sbjct 1263 EGYALMPDGRSC 1274
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 37/70 (52%), Gaps = 1/70 (1%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
+ C NT+GS++C C GY C DVDEC G C + A C+NV GSF+C+C
Sbjct 1330 MFGECENTKGSFICHCQLGYS-VKKGTTGCTDVDECEIGAHNCDMHASCLNVPGSFKCSC 1388
Query 85 MAGFEAADAK 94
G+ K
Sbjct 1389 REGWVGNGIK 1398
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 31/71 (43%), Positives = 39/71 (54%), Gaps = 3/71 (4%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C N+EGSY C+C+ GY DG +C D+DEC C QC N+ G + C C
Sbjct 1248 TQCTNSEGSYECSCSEGYA-LMPDGRSCADIDECENNPDICD-GGQCTNIPGEYRCLCYD 1305
Query 87 GFEAA-DAKTC 96
GF A+ D KTC
Sbjct 1306 GFMASMDMKTC 1316
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 39/66 (59%), Gaps = 3/66 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT+GS+ C CN G+E ++DG C D DEC T ++ C+N DGSF+C C
Sbjct 581 NGRCVNTDGSFQCICNAGFE-LTTDGKNCVDHDECT--TTNMCLNGMCINEDGSFKCVCK 637
Query 86 AGFEAA 91
GF A
Sbjct 638 PGFILA 643
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 42/73 (57%), Gaps = 5/73 (6%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ +C N GS+ CTCN G+EP C+D++ECA C + +C+N GS+ECTC
Sbjct 2218 NGTCTNVIGSFECTCNEGFEPGPM--MNCEDINECAQNPLLC--AFRCMNTFGSYECTCP 2273
Query 86 AGFEAA-DAKTCK 97
G+ D K CK
Sbjct 2274 VGYALREDQKMCK 2286
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/63 (44%), Positives = 34/63 (53%), Gaps = 2/63 (3%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N GSY C CN GYEP +S G C D+DEC C + C N GS+ CTC
Sbjct 774 NGICENLRGSYRCNCNSGYEPDAS-GRNCIDIDECLVNRLLCD-NGLCRNTPGSYSCTCP 831
Query 86 AGF 88
G+
Sbjct 832 PGY 834
Score = 51.2 bits (121), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 41/72 (56%), Gaps = 3/72 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ +C NT GSY C C PG+E + + C D+DEC++ + + +C N GSF+C C
Sbjct 1938 NGTCKNTVGSYNCLCYPGFE--LTHNNDCLDIDECSSFFGQVCRNGRCFNEIGSFKCLCN 1995
Query 86 AGFE-AADAKTC 96
G+E D K C
Sbjct 1996 EGYELTPDGKNC 2007
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/76 (38%), Positives = 37/76 (48%), Gaps = 2/76 (2%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E S+ + C N GS+ C CN GYE + DG C D +EC A C C
Sbjct 1969 ECSSFFGQVCRNGRCFNEIGSFKCLCNEGYE-LTPDGKNCIDTNECVALPGSCS-PGTCQ 2026
Query 75 NVDGSFECTCMAGFEA 90
N++GSF C C G+E
Sbjct 2027 NLEGSFRCICPPGYEV 2042
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 30/72 (41%), Positives = 36/72 (50%), Gaps = 3/72 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT+GS+ C C GY G C D DEC+ G + C NV GSFECTC
Sbjct 2177 NGQCINTDGSFRCECPMGYN-LDYTGVRCVDTDECSIGNP--CGNGTCTNVIGSFECTCN 2233
Query 86 AGFEAADAKTCK 97
GFE C+
Sbjct 2234 EGFEPGPMMNCE 2245
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 29/70 (41%), Positives = 34/70 (48%), Gaps = 3/70 (4%)
Query 19 AHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDG 78
+ S + C NT+GSY C+C GY DG CKD+DEC C CVN G
Sbjct 2489 SQSPKPCNFICKNTKGSYQCSCPRGYV-LQEDGKTCKDLDECQTKQHNCQF--LCVNTLG 2545
Query 79 SFECTCMAGF 88
F C C GF
Sbjct 2546 GFTCKCPPGF 2555
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 39/80 (48%), Gaps = 2/80 (2%)
Query 16 ASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVN 75
A H S C N G+++C C PG DG C D +EC C + +CVN
Sbjct 2292 AEGLHDCESRGMMCKNLIGTFMCICPPGMA-RRPDGEGCVDENECRTKPGICE-NGRCVN 2349
Query 76 VDGSFECTCMAGFEAADAKT 95
+ GS+ C C GF+++ + T
Sbjct 2350 IIGSYRCECNEGFQSSSSGT 2369
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 31/94 (32%), Positives = 42/94 (44%), Gaps = 4/94 (4%)
Query 1 LPMVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDEC 60
L M R C + + S + + C NT GS+ C C GYE C D+DEC
Sbjct 1099 LDMEERNCTDIDECRISPDLCGSGI---CVNTPGSFECECFEGYESGFMMMKNCMDIDEC 1155
Query 61 AAGTAECHVSAQCVNVDGSFECTCMAGFEAADAK 94
C CVN +GSF+C C G E + ++
Sbjct 1156 ERNPLLCR-GGTCVNTEGSFQCDCPLGHELSPSR 1188
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 38/73 (52%), Gaps = 4/73 (5%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
L+ C N +GS+ C C PG+ A +G C DVDEC T ++ C+N +GSF C C
Sbjct 621 LNGMCINEDGSFKCVCKPGFILA-PNGRYCTDVDECQ--TPGICMNGHCINNEGSFRCDC 677
Query 85 MAGFE-AADAKTC 96
G D + C
Sbjct 678 PPGLAVGVDGRVC 690
Score = 48.5 bits (114), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 34/66 (51%), Gaps = 2/66 (3%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT GS+ C CN G+ + C D+DEC C S CVN GSFEC C
Sbjct 1079 YGKCRNTIGSFKCRCNNGFA-LDMEERNCTDIDECRISPDLCG-SGICVNTPGSFECECF 1136
Query 86 AGFEAA 91
G+E+
Sbjct 1137 EGYESG 1142
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 31/77 (40%), Positives = 40/77 (51%), Gaps = 4/77 (5%)
Query 22 STSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFE 81
S + C NT GS+ C C GY S G AC D+DEC+ C+ C N GS++
Sbjct 2451 SLCTNGQCVNTMGSFRCFCKVGYTTDIS-GTACVDLDECSQSPKPCNFI--CKNTKGSYQ 2507
Query 82 CTCMAGFEAA-DAKTCK 97
C+C G+ D KTCK
Sbjct 2508 CSCPRGYVLQEDGKTCK 2524
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/61 (45%), Positives = 32/61 (52%), Gaps = 2/61 (3%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSA-QCVNVDGSFECTCMAG 87
C NT GSY CTC GY D CKD+DECA G +C C N+ G+F C C G
Sbjct 2261 CMNTFGSYECTCPVGYA-LREDQKMCKDLDECAEGLHDCESRGMMCKNLIGTFMCICPPG 2319
Query 88 F 88
Sbjct 2320 M 2320
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/92 (29%), Positives = 50/92 (54%), Gaps = 5/92 (5%)
Query 3 MVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAA 62
M+ + C+ + + E + +C NTEGS+ C C G+E + S C D++EC+
Sbjct 1144 MMMKNCMDIDECERNPL---LCRGGTCVNTEGSFQCDCPLGHELSPSR-EDCVDINECSL 1199
Query 63 GTAECHVSAQCVNVDGSFECTCMAGFEAADAK 94
C + +CVN+ G+++C+C G++A +
Sbjct 1200 SDNLCR-NGKCVNMIGTYQCSCNPGYQATPDR 1230
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 37/71 (52%), Gaps = 1/71 (1%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N GS+ C C G+ + C+D+DEC+ G C +A C+N GS+ C C
Sbjct 1814 NGVCINQIGSFRCECPTGFS-YNDLLLVCEDIDECSNGDNLCQRNADCINSPGSYRCECA 1872
Query 86 AGFEAADAKTC 96
AGF+ + C
Sbjct 1873 AGFKLSPNGAC 1883
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/71 (39%), Positives = 34/71 (47%), Gaps = 4/71 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H C + +GSY C CN G++ AS D C DVDEC + C N GS+ C C
Sbjct 1898 HGLCVDLQGSYQCICNNGFK-ASQDQTMCMDVDECERHPCG---NGTCKNTVGSYNCLCY 1953
Query 86 AGFEAADAKTC 96
GFE C
Sbjct 1954 PGFELTHNNDC 1964
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 34/66 (51%), Gaps = 4/66 (6%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
+C N EGS+ C C PGYE S + C D++EC C + C N G F+C C
Sbjct 2023 GTCQNLEGSFRCICPPGYEVRSEN---CIDINECDEDPNIC-LFGSCTNTPGGFQCICPP 2078
Query 87 GFEAAD 92
GF +D
Sbjct 2079 GFVLSD 2084
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/76 (39%), Positives = 39/76 (51%), Gaps = 7/76 (9%)
Query 20 HSSTSLHASCANTEGSYVCTCNPGY-EPASSDGHACKDVDECAAGTAECHVSAQCVNVDG 78
H++ L+ C T SY C CN GY + A+ D C DVDEC + + CVN G
Sbjct 494 HANLCLNGRCIPTVSSYRCECNMGYKQDANGD---CIDVDECTSNPCS---NGDCVNTPG 547
Query 79 SFECTCMAGFEAADAK 94
S+ C C AGF+ K
Sbjct 548 SYYCKCHAGFQRTPTK 563
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 26/72 (36%), Positives = 38/72 (52%), Gaps = 3/72 (4%)
Query 18 AAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVD 77
A +++ + C N G+Y C C G+ PA SD +C+D+DEC+ V C N+
Sbjct 1447 AENTNLCENGQCLNVPGAYRCECEMGFTPA-SDSRSCQDIDECSFQNI--CVFGTCNNLP 1503
Query 78 GSFECTCMAGFE 89
G F C C G+E
Sbjct 1504 GMFHCICDDGYE 1515
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 41/81 (50%), Gaps = 3/81 (3%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E S + +A C N+ GSY C C G++ S AC D +EC C CV
Sbjct 1846 ECSNGDNLCQRNADCINSPGSYRCECAAGFK--LSPNGACVDRNECLEIPNVCS-HGLCV 1902
Query 75 NVDGSFECTCMAGFEAADAKT 95
++ GS++C C GF+A+ +T
Sbjct 1903 DLQGSYQCICNNGFKASQDQT 1923
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 35/65 (53%), Gaps = 3/65 (4%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
+ +C N G + C C+ GYE + G+ C D+DECA C V+ CVN G +EC C
Sbjct 1495 VFGTCNNLPGMFHCICDDGYELDRTGGN-CTDIDECAD-PINC-VNGLCVNTPGRYECNC 1551
Query 85 MAGFE 89
F+
Sbjct 1552 PPDFQ 1556
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 29/60 (48%), Gaps = 3/60 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT G + C C PG+ + AC D +EC + + C C N GSF C C GF
Sbjct 2540 CVNTLGGFTCKCPPGFTQHHT---ACIDNNECGSQPSLCGAKGICQNTPGSFSCECQRGF 2596
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 30/69 (43%), Gaps = 2/69 (2%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C N G Y C C G+ AS D C DV+EC C + +C N GSF C C
Sbjct 1290 GQCTNIPGEYRCLCYDGFM-ASMDMKTCIDVNECDLNPNIC-MFGECENTKGSFICHCQL 1347
Query 87 GFEAADAKT 95
G+ T
Sbjct 1348 GYSVKKGTT 1356
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 29/54 (53%), Gaps = 2/54 (3%)
Query 40 CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADA 93
C G+ P G AC+DVDEC A C C+N GSFEC C AG + ++
Sbjct 261 CRRGFIPNIRTG-ACQDVDECQAIPGLCQ-GGNCINTVGSFECRCPAGHKQSET 312
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
+C NT GS+ C C G++ S C+D+DEC+ C + C N GS+ C C
Sbjct 290 GNCINTVGSFECRCPAGHK-QSETTQKCEDIDECSVIPGVCE-TGDCSNTVGSYFCLCPR 347
Query 87 GF 88
GF
Sbjct 348 GF 349
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 32/62 (51%), Gaps = 3/62 (4%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
ASC NT GSY C C G+ AC DV+EC++ C S C N +G + C C
Sbjct 2660 ASCYNTLGSYKCACPSGFS-FDQFSSACHDVNECSSSKNPC--SYGCSNTEGGYLCGCPP 2716
Query 87 GF 88
G+
Sbjct 2717 GY 2718
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 35/78 (44%), Gaps = 4/78 (5%)
Query 11 VPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVS 70
+ E + S C NT GS+ C C G+ +S G C+DVDEC G C
Sbjct 2563 IDNNECGSQPSLCGAKGICQNTPGSFSCECQRGFSLDAS-GLNCEDVDEC-DGNHRCQHG 2620
Query 71 AQCVNVDGSFECTCMAGF 88
Q N+ G + C C G+
Sbjct 2621 CQ--NILGGYRCGCPQGY 2636
Score = 35.8 bits (81), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 29/58 (50%), Gaps = 2/58 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
+C NT GS+ C C GY S + C+D+DEC A C C N G++ C C
Sbjct 1657 GNCINTFGSFQCECPQGYY-LSEETRICEDIDECFAHPGVCG-PGTCYNTLGNYTCIC 1712
Score = 35.4 bits (80), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 29/52 (55%), Gaps = 7/52 (13%)
Query 37 VCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
+C PGY ++DG +D+DEC + C + QCVN GSF C C G+
Sbjct 2429 ICPHGPGY---ATDG---RDIDECKVMPSLC-TNGQCVNTMGSFRCFCKVGY 2473
Score = 31.6 bits (70), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 29/60 (48%), Gaps = 3/60 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N G Y C C GY + C D +EC+ A SA C N GS++C C +GF
Sbjct 2621 CQNILGGYRCGCPQGYV-QHYQWNQCVDENECSNPGA--CGSASCYNTLGSYKCACPSGF 2677
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 21/42 (50%), Gaps = 1/42 (2%)
Query 47 ASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
A G C+DV+EC C + +CVN GSF C C G
Sbjct 939 ARIKGVTCEDVNECEVFPGVCP-NGRCVNSKGSFHCECPEGL 979
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 19/39 (48%), Gaps = 1/39 (2%)
Query 19 AHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDV 57
AH +C NT G+Y C C P Y + GH C D+
Sbjct 1691 AHPGVCGPGTCYNTLGNYTCICPPEYMQVNG-GHNCMDM 1728
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 20/38 (52%), Gaps = 1/38 (2%)
Query 55 KDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAAD 92
+D+DEC C C+N GSF+C C G+ ++
Sbjct 1642 EDIDECQELPGLCQ-GGNCINTFGSFQCECPQGYYLSE 1678
Score = 28.5 bits (62), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 18/35 (51%), Gaps = 1/35 (2%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDE 59
L SC NT G + C C PG+ S +G C D +
Sbjct 2061 LFGSCTNTPGGFQCICPPGFV-LSDNGRRCFDTRQ 2094
> mmu:100047082 fibrillin-2-like
Length=2907
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 34/72 (47%), Positives = 44/72 (61%), Gaps = 4/72 (5%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
+HASC N GS+ C+C G+ +G C D+DECA GT +C ++AQCVN GS+ C C
Sbjct 1373 MHASCLNVPGSFKCSCREGW---VGNGIKCIDLDECANGTHQCSINAQCVNTPGSYRCAC 1429
Query 85 MAGFEAADAKTC 96
GF D TC
Sbjct 1430 SEGF-TGDGFTC 1440
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 34/75 (45%), Positives = 48/75 (64%), Gaps = 5/75 (6%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
S++A C NT GSY C C+ G+ + DG C DVDECA T C + QC+NV G++ C
Sbjct 1413 SINAQCVNTPGSYRCACSEGF---TGDGFTCSDVDECAENTNLCE-NGQCLNVPGAYRCE 1468
Query 84 CMAGF-EAADAKTCK 97
C GF A+D+++C+
Sbjct 1469 CEMGFTPASDSRSCQ 1483
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 41/72 (56%), Gaps = 3/72 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT GSY C C+ G++ + AC D+DEC C + +CVN DGSF+C C
Sbjct 539 NGDCVNTPGSYYCKCHAGFQRTPTK-QACIDIDECIQNGVLCK-NGRCVNSDGSFQCICN 596
Query 86 AGFE-AADAKTC 96
AGFE D K C
Sbjct 597 AGFELTTDGKNC 608
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 41/72 (56%), Gaps = 4/72 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N G+Y C+CNPGY+ A+ D C D+DEC C QC N +GS+EC+C
Sbjct 1206 NGKCVNMIGTYQCSCNPGYQ-ATPDRQGCTDIDECMIMNGGC--DTQCTNSEGSYECSCS 1262
Query 86 AGFE-AADAKTC 96
G+ D ++C
Sbjct 1263 EGYALMPDGRSC 1274
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 37/70 (52%), Gaps = 1/70 (1%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
+ C NT+GS++C C GY C DVDEC G C + A C+NV GSF+C+C
Sbjct 1330 MFGECENTKGSFICHCQLGYS-VKKGTTGCTDVDECEIGAHNCDMHASCLNVPGSFKCSC 1388
Query 85 MAGFEAADAK 94
G+ K
Sbjct 1389 REGWVGNGIK 1398
Score = 54.7 bits (130), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 31/71 (43%), Positives = 39/71 (54%), Gaps = 3/71 (4%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C N+EGSY C+C+ GY DG +C D+DEC C QC N+ G + C C
Sbjct 1248 TQCTNSEGSYECSCSEGYA-LMPDGRSCADIDECENNPDICD-GGQCTNIPGEYRCLCYD 1305
Query 87 GFEAA-DAKTC 96
GF A+ D KTC
Sbjct 1306 GFMASMDMKTC 1316
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/66 (42%), Positives = 39/66 (59%), Gaps = 3/66 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N++GS+ C CN G+E ++DG C D DEC T ++ C+N DGSF+C C
Sbjct 581 NGRCVNSDGSFQCICNAGFE-LTTDGKNCVDHDECT--TTNMCLNGMCINEDGSFKCVCK 637
Query 86 AGFEAA 91
GF A
Sbjct 638 PGFILA 643
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 27/63 (42%), Positives = 34/63 (53%), Gaps = 2/63 (3%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N G+Y C CN GYEP +S G C D+DEC C + C N GS+ CTC
Sbjct 774 NGICENLRGTYRCNCNSGYEPDAS-GRNCIDIDECLVNRLLCD-NGLCRNTPGSYSCTCP 831
Query 86 AGF 88
G+
Sbjct 832 PGY 834
Score = 51.2 bits (121), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 41/72 (56%), Gaps = 3/72 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ +C NT GSY C C PG+E + + C D+DEC++ + + +C N GSF+C C
Sbjct 1938 NGTCKNTVGSYNCLCYPGFE--LTHNNDCLDIDECSSFFGQVCRNGRCFNEIGSFKCLCN 1995
Query 86 AGFE-AADAKTC 96
G+E D K C
Sbjct 1996 EGYELTPDGKNC 2007
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/76 (38%), Positives = 37/76 (48%), Gaps = 2/76 (2%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E S+ + C N GS+ C CN GYE + DG C D +EC A C C
Sbjct 1969 ECSSFFGQVCRNGRCFNEIGSFKCLCNEGYE-LTPDGKNCIDTNECVALPGSCS-PGTCQ 2026
Query 75 NVDGSFECTCMAGFEA 90
N++GSF C C G+E
Sbjct 2027 NLEGSFRCICPPGYEV 2042
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 41/73 (56%), Gaps = 5/73 (6%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ +C N G + CTCN G+EP C+D++ECA C + +C+N GS+ECTC
Sbjct 2218 NGTCTNVIGCFECTCNEGFEPGPM--MNCEDINECAQNPLLC--AFRCMNTFGSYECTCP 2273
Query 86 AGFEAA-DAKTCK 97
G+ D K CK
Sbjct 2274 VGYGLREDQKMCK 2286
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 39/80 (48%), Gaps = 2/80 (2%)
Query 16 ASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVN 75
A H S C N G+++C C PG DG C D +EC C + +CVN
Sbjct 2292 AEGLHDCESRGMMCKNLIGTFMCICPPGMA-RRPDGEGCVDENECRTKPGICE-NGRCVN 2349
Query 76 VDGSFECTCMAGFEAADAKT 95
+ GS+ C C GF+++ + T
Sbjct 2350 IIGSYRCECNEGFQSSSSGT 2369
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 29/70 (41%), Positives = 34/70 (48%), Gaps = 3/70 (4%)
Query 19 AHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDG 78
+ S + C NT+GSY C+C GY DG CKD+DEC C CVN G
Sbjct 2489 SQSPKPCNFICKNTKGSYQCSCPRGYV-LQEDGKTCKDLDECQTKQHNCQF--LCVNTLG 2545
Query 79 SFECTCMAGF 88
F C C GF
Sbjct 2546 GFTCKCPPGF 2555
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 31/94 (32%), Positives = 42/94 (44%), Gaps = 4/94 (4%)
Query 1 LPMVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDEC 60
L M R C + + S + + C NT GS+ C C GYE C D+DEC
Sbjct 1099 LDMEERNCTDIDECRISPDLCGSGI---CVNTPGSFECECFEGYESGFMMMKNCMDIDEC 1155
Query 61 AAGTAECHVSAQCVNVDGSFECTCMAGFEAADAK 94
C CVN +GSF+C C G E + ++
Sbjct 1156 ERNPLLCR-GGTCVNTEGSFQCDCPLGHELSPSR 1188
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 38/73 (52%), Gaps = 4/73 (5%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
L+ C N +GS+ C C PG+ A +G C DVDEC T ++ C+N +GSF C C
Sbjct 621 LNGMCINEDGSFKCVCKPGFILA-PNGRYCTDVDECQ--TPGICMNGHCINNEGSFRCDC 677
Query 85 MAGFE-AADAKTC 96
G D + C
Sbjct 678 PPGLAVGVDGRVC 690
Score = 48.1 bits (113), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 34/66 (51%), Gaps = 2/66 (3%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT GS+ C CN G+ + C D+DEC C S CVN GSFEC C
Sbjct 1079 YGKCRNTIGSFKCRCNNGFA-LDMEERNCTDIDECRISPDLCG-SGICVNTPGSFECECF 1136
Query 86 AGFEAA 91
G+E+
Sbjct 1137 EGYESG 1142
Score = 48.1 bits (113), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 35/72 (48%), Gaps = 3/72 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT+GS+ C C GY G C D DEC+ G + C NV G FECTC
Sbjct 2177 NGQCINTDGSFRCECPMGYN-LDYTGVRCVDTDECSIGNP--CGNGTCTNVIGCFECTCN 2233
Query 86 AGFEAADAKTCK 97
GFE C+
Sbjct 2234 EGFEPGPMMNCE 2245
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 28/71 (39%), Positives = 34/71 (47%), Gaps = 4/71 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H C + +GSY C CN G++ AS D C DVDEC + C N GS+ C C
Sbjct 1898 HGLCVDLQGSYQCICNNGFK-ASQDQTMCMDVDECERHPC---ANGTCKNTVGSYNCLCY 1953
Query 86 AGFEAADAKTC 96
GFE C
Sbjct 1954 PGFELTHNNDC 1964
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 31/77 (40%), Positives = 40/77 (51%), Gaps = 4/77 (5%)
Query 22 STSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFE 81
S + C NT GS+ C C GY S G AC D+DEC+ C+ C N GS++
Sbjct 2451 SLCTNGQCVNTMGSFRCFCKVGYTMDIS-GTACVDLDECSQSPKPCNFI--CKNTKGSYQ 2507
Query 82 CTCMAGFEAA-DAKTCK 97
C+C G+ D KTCK
Sbjct 2508 CSCPRGYVLQEDGKTCK 2524
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/61 (45%), Positives = 32/61 (52%), Gaps = 2/61 (3%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSA-QCVNVDGSFECTCMAG 87
C NT GSY CTC GY D CKD+DECA G +C C N+ G+F C C G
Sbjct 2261 CMNTFGSYECTCPVGYG-LREDQKMCKDLDECAEGLHDCESRGMMCKNLIGTFMCICPPG 2319
Query 88 F 88
Sbjct 2320 M 2320
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/92 (29%), Positives = 50/92 (54%), Gaps = 5/92 (5%)
Query 3 MVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAA 62
M+ + C+ + + E + +C NTEGS+ C C G+E + S C D++EC+
Sbjct 1144 MMMKNCMDIDECERNPL---LCRGGTCVNTEGSFQCDCPLGHELSPSR-EDCVDINECSL 1199
Query 63 GTAECHVSAQCVNVDGSFECTCMAGFEAADAK 94
C + +CVN+ G+++C+C G++A +
Sbjct 1200 SDNLCR-NGKCVNMIGTYQCSCNPGYQATPDR 1230
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 37/71 (52%), Gaps = 1/71 (1%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N GS+ C C G+ + C+D+DEC+ G C +A C+N GS+ C C
Sbjct 1814 NGVCINQIGSFRCECPTGFS-YNDLLLVCEDIDECSNGDNLCQRNADCINSPGSYRCECA 1872
Query 86 AGFEAADAKTC 96
AGF+ + C
Sbjct 1873 AGFKLSPNGAC 1883
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 34/66 (51%), Gaps = 4/66 (6%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
+C N EGS+ C C PGYE S + C D++EC C + C N G F+C C
Sbjct 2023 GTCQNLEGSFRCICPPGYEVRSEN---CIDINECDEDPNIC-LFGSCTNTPGGFQCICPP 2078
Query 87 GFEAAD 92
GF +D
Sbjct 2079 GFVLSD 2084
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/76 (39%), Positives = 39/76 (51%), Gaps = 7/76 (9%)
Query 20 HSSTSLHASCANTEGSYVCTCNPGY-EPASSDGHACKDVDECAAGTAECHVSAQCVNVDG 78
H++ L+ C T SY C CN GY + A+ D C DVDEC + + CVN G
Sbjct 494 HANLCLNGRCIPTVSSYRCRCNMGYKQDANGD---CIDVDECTSNPCS---NGDCVNTPG 547
Query 79 SFECTCMAGFEAADAK 94
S+ C C AGF+ K
Sbjct 548 SYYCKCHAGFQRTPTK 563
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 26/72 (36%), Positives = 38/72 (52%), Gaps = 3/72 (4%)
Query 18 AAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVD 77
A +++ + C N G+Y C C G+ PA SD +C+D+DEC+ V C N+
Sbjct 1447 AENTNLCENGQCLNVPGAYRCECEMGFTPA-SDSRSCQDIDECSFQNI--CVFGTCNNLP 1503
Query 78 GSFECTCMAGFE 89
G F C C G+E
Sbjct 1504 GMFHCICDDGYE 1515
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 43/81 (53%), Gaps = 3/81 (3%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E S + +A C N+ GSY C C G++ S +G AC D +EC C CV
Sbjct 1846 ECSNGDNLCQRNADCINSPGSYRCECAAGFK-LSPNG-ACVDRNECLEIPNVCS-HGLCV 1902
Query 75 NVDGSFECTCMAGFEAADAKT 95
++ GS++C C GF+A+ +T
Sbjct 1903 DLQGSYQCICNNGFKASQDQT 1923
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 35/65 (53%), Gaps = 3/65 (4%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
+ +C N G + C C+ GYE + G+ C D+DECA C V+ CVN G +EC C
Sbjct 1495 VFGTCNNLPGMFHCICDDGYELDRTGGN-CTDIDECAD-PINC-VNGLCVNTPGRYECNC 1551
Query 85 MAGFE 89
F+
Sbjct 1552 PPDFQ 1556
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 24/54 (44%), Positives = 30/54 (55%), Gaps = 2/54 (3%)
Query 40 CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADA 93
C PG+ P G AC+DVDEC A C C+N GSFEC C AG + ++
Sbjct 261 CRPGFIPNIRTG-ACQDVDECQAIPGLCQ-GGNCINTVGSFECRCPAGHKQSET 312
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 29/60 (48%), Gaps = 3/60 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT G + C C PG+ + AC D +EC + + C C N GSF C C GF
Sbjct 2540 CVNTLGGFTCKCPPGFTQHHT---ACIDNNECGSQPSLCGAKGICQNTPGSFSCECQRGF 2596
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 30/69 (43%), Gaps = 2/69 (2%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C N G Y C C G+ AS D C DV+EC C + +C N GSF C C
Sbjct 1290 GQCTNIPGEYRCLCYDGFM-ASMDMKTCIDVNECDLNPNIC-MFGECENTKGSFICHCQL 1347
Query 87 GFEAADAKT 95
G+ T
Sbjct 1348 GYSVKKGTT 1356
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
+C NT GS+ C C G++ S C+D+DEC+ C + C N GS+ C C
Sbjct 290 GNCINTVGSFECRCPAGHK-QSETTQKCEDIDECSVIPGVCE-TGDCSNTVGSYFCLCPR 347
Query 87 GF 88
GF
Sbjct 348 GF 349
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 32/62 (51%), Gaps = 3/62 (4%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
ASC NT GSY C C G+ AC DV+EC++ C S C N +G + C C
Sbjct 2660 ASCYNTLGSYKCACPSGFS-FDQFSSACHDVNECSSSKNPC--SYGCSNTEGGYLCGCPP 2716
Query 87 GF 88
G+
Sbjct 2717 GY 2718
Score = 35.8 bits (81), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 29/58 (50%), Gaps = 2/58 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
+C NT GS+ C C GY S + C+D+DEC A C C N G++ C C
Sbjct 1657 GNCINTFGSFQCECPQGYY-LSEETRICEDIDECFAHPGVCG-PGTCYNTLGNYTCIC 1712
Score = 35.4 bits (80), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 34/77 (44%), Gaps = 4/77 (5%)
Query 11 VPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVS 70
+ E + S C NT GS+ C C G+ +S G C+DVDEC G C
Sbjct 2563 IDNNECGSQPSLCGAKGICQNTPGSFSCECQRGFSLDAS-GLNCEDVDEC-DGNHRCQHG 2620
Query 71 AQCVNVDGSFECTCMAG 87
Q N+ G + C C G
Sbjct 2621 CQ--NILGGYRCGCPHG 2635
Score = 35.4 bits (80), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 29/52 (55%), Gaps = 7/52 (13%)
Query 37 VCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
+C PGY ++DG +D+DEC + C + QCVN GSF C C G+
Sbjct 2429 ICPHGPGY---ATDG---RDIDECKVMPSLC-TNGQCVNTMGSFRCFCKVGY 2473
Score = 32.0 bits (71), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 20/35 (57%), Gaps = 1/35 (2%)
Query 54 CKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
CKD++EC A C +C N GSF+C C GF
Sbjct 1064 CKDINECKAFPGMC-TYGKCRNTIGSFKCRCNNGF 1097
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 21/42 (50%), Gaps = 1/42 (2%)
Query 47 ASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
A G C+DV+EC C + +CVN GSF C C G
Sbjct 939 ARIKGVTCEDVNECEVFPGVCP-NGRCVNSKGSFHCECPEGL 979
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 29/60 (48%), Gaps = 3/60 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N G Y C C P + + C D +EC+ A SA C N GS++C C +GF
Sbjct 2621 CQNILGGYRCGC-PHGDVQHYQWNQCVDENECSNPGA--CGSASCYNTLGSYKCACPSGF 2677
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 19/39 (48%), Gaps = 1/39 (2%)
Query 19 AHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDV 57
AH +C NT G+Y C C P Y + GH C D+
Sbjct 1691 AHPGVCGPGTCYNTLGNYTCICPPEYMQVNG-GHNCMDM 1728
Score = 28.5 bits (62), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 20/38 (52%), Gaps = 1/38 (2%)
Query 55 KDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAAD 92
+D+DEC C C+N GSF+C C G+ ++
Sbjct 1642 EDIDECQELPGLCQ-GGNCINTFGSFQCECPQGYYLSE 1678
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 18/35 (51%), Gaps = 1/35 (2%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDE 59
L SC NT G + C C PG+ S +G C D +
Sbjct 2061 LFGSCTNTPGGFQCICPPGFV-LSDNGRRCFDTRQ 2094
> hsa:84467 FBN3, KIAA1776; fibrillin 3
Length=2809
Score = 64.7 bits (156), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 39/94 (41%), Positives = 52/94 (55%), Gaps = 8/94 (8%)
Query 4 VFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAG 63
V +C+ V + +S H H C N G+Y C C PG++ A+ AC DVDEC
Sbjct 443 VRGECIDVDECTSSPCH-----HGDCVNIPGTYHCRCYPGFQ-ATPTRQACVDVDECIVS 496
Query 64 TAECHVSAQCVNVDGSFECTCMAGFE-AADAKTC 96
CH+ +CVN +GSF+C C AGFE + D K C
Sbjct 497 GGLCHL-GRCVNTEGSFQCVCNAGFELSPDGKNC 529
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 35/92 (38%), Positives = 53/92 (57%), Gaps = 5/92 (5%)
Query 3 MVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAA 62
M+ + C+ V + A +C NT+GSY C C PG+E ++ G AC+D+DEC+
Sbjct 1064 MLMKNCMDV---DECARDPLLCRGGTCTNTDGSYKCQCPPGHE-LTAKGTACEDIDECSL 1119
Query 63 GTAECHVSAQCVNVDGSFECTCMAGFEAADAK 94
C QCVNV G+F+C+C AGF++ +
Sbjct 1120 SDGLC-PHGQCVNVIGAFQCSCHAGFQSTPDR 1150
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 39/72 (54%), Gaps = 4/72 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
HASC N GS+ C C PG+ DG C D+DEC + C C+NV GS+ CTC
Sbjct 1294 HASCLNIPGSFSCRCLPGW---VGDGFECHDLDECVSQEHRCSPRGDCLNVPGSYRCTCR 1350
Query 86 AGFEAADAKTCK 97
GF A D C+
Sbjct 1351 QGF-AGDGFFCE 1361
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 29/64 (45%), Positives = 35/64 (54%), Gaps = 1/64 (1%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
LH C NT+GS+VC C GY C DVDEC G C A C+N+ GSF C C
Sbjct 1250 LHGDCENTKGSFVCHCQLGYM-VRKGATGCSDVDECEVGGHNCDSHASCLNIPGSFSCRC 1308
Query 85 MAGF 88
+ G+
Sbjct 1309 LPGW 1312
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 31/62 (50%), Positives = 37/62 (59%), Gaps = 2/62 (3%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSA-QCVNVDGSFECTCMAG 87
C NTEGSY+CTC GY DG C+DVDECA G +CH +C N+ G+F C C G
Sbjct 2181 CHNTEGSYLCTCPAGYT-LREDGAMCRDVDECADGQQDCHARGMECKNLIGTFACVCPPG 2239
Query 88 FE 89
Sbjct 2240 MR 2241
Score = 54.7 bits (130), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 32/65 (49%), Positives = 37/65 (56%), Gaps = 3/65 (4%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C NTEGS+ C CN G+E S DG C D +ECA T V+ C+N DGSF C C
Sbjct 503 GRCVNTEGSFQCVCNAGFE-LSPDGKNCVDHNECATSTM--CVNGVCLNEDGSFSCLCKP 559
Query 87 GFEAA 91
GF A
Sbjct 560 GFLLA 564
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 41/79 (51%), Gaps = 4/79 (5%)
Query 19 AHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDG 78
A S+ ++ C N +GS+ C C PG+ A GH C D+DEC T V+ C N +G
Sbjct 536 ATSTMCVNGVCLNEDGSFSCLCKPGFLLAPG-GHYCMDIDECQ--TPGICVNGHCTNTEG 592
Query 79 SFECTCMAGFEAA-DAKTC 96
SF C C+ G D + C
Sbjct 593 SFRCQCLGGLAVGTDGRVC 611
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 32/73 (43%), Positives = 39/73 (53%), Gaps = 3/73 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N GSY C CN GYE A + G C DVDECA + C + C N GS+ C+C
Sbjct 695 NGVCENLRGSYRCVCNLGYE-AGASGKDCTDVDECALNSLLCD-NGWCQNSPGSYSCSCP 752
Query 86 AGFE-AADAKTCK 97
GF D + CK
Sbjct 753 PGFHFWQDTEICK 765
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 32/69 (46%), Positives = 35/69 (50%), Gaps = 3/69 (4%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NTEGSY C+C GY DG AC DVDEC C C N+ G C C GF
Sbjct 1170 CINTEGSYRCSCGQGYS-LMPDGRACADVDECEENPRVCD-QGHCTNMPGGHRCLCYDGF 1227
Query 89 EAA-DAKTC 96
A D +TC
Sbjct 1228 MATPDMRTC 1236
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 34/67 (50%), Gaps = 1/67 (1%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N G++ C C PG P G C D +EC A C V+ +CVN GSF C C GF
Sbjct 2225 CKNLIGTFACVCPPGMRPLPGSGEGCTDDNECHAQPDLC-VNGRCVNTAGSFRCDCDEGF 2283
Query 89 EAADAKT 95
+ + T
Sbjct 2284 QPSPTLT 2290
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 29/70 (41%), Positives = 33/70 (47%), Gaps = 1/70 (1%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+C NT GS+ C C PGYE C DVDECA C C N DGS++C C
Sbjct 1041 QGTCVNTPGSFECECFPGYESGFMLMKNCMDVDECARDPLLCR-GGTCTNTDGSYKCQCP 1099
Query 86 AGFEAADAKT 95
G E T
Sbjct 1100 PGHELTAKGT 1109
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/60 (45%), Positives = 34/60 (56%), Gaps = 3/60 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT+GS++C+C GY DG CKD+DEC + C CVN G+F C C GF
Sbjct 2420 CKNTKGSFLCSCPRGYL-LEEDGRTCKDLDECTSRQHNCQF--LCVNTVGAFTCRCPPGF 2476
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/75 (38%), Positives = 38/75 (50%), Gaps = 5/75 (6%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
S C N GSY CTC G+ + DG C+D DECA C + QC+N G + C
Sbjct 1333 SPRGDCLNVPGSYRCTCRQGF---AGDGFFCEDRDECAENVDLCD-NGQCLNAPGGYRCE 1388
Query 84 CMAGFEAA-DAKTCK 97
C GF+ D + C+
Sbjct 1389 CEMGFDPTEDHRACQ 1403
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/68 (36%), Positives = 38/68 (55%), Gaps = 1/68 (1%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N GS+ C C G+ +S AC+DVDEC + + C +A C+N+ GS+ C C G+
Sbjct 1737 CINQIGSFRCECPAGFN-YNSILLACEDVDECGSRESPCQQNADCINIPGSYRCKCTRGY 1795
Query 89 EAADAKTC 96
+ + C
Sbjct 1796 KLSPGGAC 1803
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 38/72 (52%), Gaps = 4/72 (5%)
Query 21 SSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSF 80
+ T L +C N EGS+ C C PG++ S C D+DEC+ C + C N GSF
Sbjct 1937 AGTCLPGTCQNLEGSFRCICPPGFQVQSDH---CIDIDECSEEPNLC-LFGTCTNSPGSF 1992
Query 81 ECTCMAGFEAAD 92
+C C GF +D
Sbjct 1993 QCLCPPGFVLSD 2004
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 26/63 (41%), Positives = 36/63 (57%), Gaps = 4/63 (6%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H C +TEGSY+C C+ G++ AS+D C D+DEC + C N+ GS+ C C
Sbjct 1818 HGDCMDTEGSYMCLCHRGFQ-ASADQTLCMDIDECDRQPCG---NGTCKNIIGSYNCLCF 1873
Query 86 AGF 88
GF
Sbjct 1874 PGF 1876
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 33/64 (51%), Gaps = 3/64 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N G Y C C G++P + D AC+DVDECA G C N+ G F C C
Sbjct 1375 NGQCLNAPGGYRCECEMGFDP-TEDHRACQDVDECAQGNL--CAFGSCENLPGMFRCICN 1431
Query 86 AGFE 89
G+E
Sbjct 1432 GGYE 1435
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 35/62 (56%), Gaps = 2/62 (3%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT GS+ C C G+E ++DG C D +EC + C + C N++GSF C C GF
Sbjct 1903 CLNTAGSFHCLCQDGFE-LTADGKNCVDTNECLSLAGTC-LPGTCQNLEGSFRCICPPGF 1960
Query 89 EA 90
+
Sbjct 1961 QV 1962
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 32/82 (39%), Positives = 38/82 (46%), Gaps = 7/82 (8%)
Query 18 AAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQ--CVN 75
A + + C NT+GS+ C C GY G C D DEC+ G H Q C N
Sbjct 2089 AENPGVCTNGVCVNTDGSFRCECPFGYS-LDFTGINCVDTDECSVG----HPCGQGTCTN 2143
Query 76 VDGSFECTCMAGFEAADAKTCK 97
V G FEC C GFE TC+
Sbjct 2144 VIGGFECACADGFEPGLMMTCE 2165
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 34/72 (47%), Gaps = 3/72 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ +C N GSY C C PG+ + C D DEC + C+N GSF C C
Sbjct 1858 NGTCKNIIGSYNCLCFPGF--VVTHNGDCVDFDECTTLVGQVCRFGHCLNTAGSFHCLCQ 1915
Query 86 AGFE-AADAKTC 96
GFE AD K C
Sbjct 1916 DGFELTADGKNC 1927
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 39/72 (54%), Gaps = 4/72 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H C N G++ C+C+ G++ ++ D C D++EC C V C+N +GS+ C+C
Sbjct 1126 HGQCVNVIGAFQCSCHAGFQ-STPDRQGCVDINECRVQNGGCDV--HCINTEGSYRCSCG 1182
Query 86 AGFE-AADAKTC 96
G+ D + C
Sbjct 1183 QGYSLMPDGRAC 1194
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 34/66 (51%), Gaps = 2/66 (3%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H +C NT GS+ C C G+ + + C D+DEC C CVN GSFEC C
Sbjct 999 HGTCRNTVGSFHCACAGGFALDAQE-RNCTDIDECRISPDLCG-QGTCVNTPGSFECECF 1056
Query 86 AGFEAA 91
G+E+
Sbjct 1057 PGYESG 1062
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/60 (41%), Positives = 30/60 (50%), Gaps = 3/60 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT G++ C C PG+ + AC D DEC+A C C N GSF C C GF
Sbjct 2461 CVNTVGAFTCRCPPGF---TQHHQACFDNDECSAQPGPCGAHGHCHNTPGSFRCECHQGF 2517
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 38/73 (52%), Gaps = 4/73 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H C N+ GS+ C C GY P ++ C D+DEC+ C + C N GSF C+C
Sbjct 2376 HGECINSLGSFRCHCQAGYTPDAT-ATTCLDMDECSQVPKPC--TFLCKNTKGSFLCSCP 2432
Query 86 AGFEA-ADAKTCK 97
G+ D +TCK
Sbjct 2433 RGYLLEEDGRTCK 2445
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 30/74 (40%), Positives = 37/74 (50%), Gaps = 4/74 (5%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E SA H C NT GS+ C C+ G+ SS GH C+DV+EC G C Q
Sbjct 2488 ECSAQPGPCGAHGHCHNTPGSFRCECHQGFTLVSS-GHGCEDVNEC-DGPHRCQHGCQ-- 2543
Query 75 NVDGSFECTCMAGF 88
N G + C+C GF
Sbjct 2544 NQLGGYRCSCPQGF 2557
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 36/70 (51%), Gaps = 4/70 (5%)
Query 18 AAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVD 77
A +S + C N+ GSY C+C PG+ D CKDVDEC + VS C N+
Sbjct 729 ALNSLLCDNGWCQNSPGSYSCSCPPGFH-FWQDTEICKDVDECLSSPC---VSGVCRNLA 784
Query 78 GSFECTCMAG 87
GS+ C C G
Sbjct 785 GSYTCKCGPG 794
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 36/71 (50%), Gaps = 3/71 (4%)
Query 19 AHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDG 78
A + SC N G + C CN GYE G+ C D++ECA C ++ C+N G
Sbjct 1409 AQGNLCAFGSCENLPGMFRCICNGGYELDRGGGN-CTDINECAD-PVNC-INGVCINTPG 1465
Query 79 SFECTCMAGFE 89
S+ C+C FE
Sbjct 1466 SYLCSCPQDFE 1476
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 37/73 (50%), Gaps = 5/73 (6%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+C N G + C C G+EP C+D+DEC+ C + +C N +GS+ CTC
Sbjct 2138 QGTCTNVIGGFECACADGFEPGLM--MTCEDIDECSLNPLLC--AFRCHNTEGSYLCTCP 2193
Query 86 AGFEAA-DAKTCK 97
AG+ D C+
Sbjct 2194 AGYTLREDGAMCR 2206
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 39/81 (48%), Gaps = 3/81 (3%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E + S +A C N GSY C C GY+ S G AC +EC C C+
Sbjct 1766 ECGSRESPCQQNADCINIPGSYRCKCTRGYK--LSPGGACVGRNECREIPNVCS-HGDCM 1822
Query 75 NVDGSFECTCMAGFEAADAKT 95
+ +GS+ C C GF+A+ +T
Sbjct 1823 DTEGSYMCLCHRGFQASADQT 1843
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 27/70 (38%), Positives = 34/70 (48%), Gaps = 3/70 (4%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
ASC NT G + C C G++ + G C++VDECA C S C N G F C C
Sbjct 2582 ASCRNTLGGFRCVCPSGFDFDQALG-GCQEVDECAGRRGPCSYS--CANTPGGFLCGCPQ 2638
Query 87 GFEAADAKTC 96
G+ A C
Sbjct 2639 GYFRAGQGHC 2648
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 31/62 (50%), Gaps = 2/62 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C NT GS+ C C PGY S C+D+DEC+ + C C N G++ C C A
Sbjct 1577 GDCVNTFGSFQCECPPGYH-LSEHTRICEDIDECSTHSGICG-PGTCYNTLGNYTCVCPA 1634
Query 87 GF 88
+
Sbjct 1635 EY 1636
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 29/70 (41%), Gaps = 2/70 (2%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
C N G + C C G+ A+ D C DVDEC C + C N GSF C C
Sbjct 1209 QGHCTNMPGGHRCLCYDGFM-ATPDMRTCVDVDECDLNPHIC-LHGDCENTKGSFVCHCQ 1266
Query 86 AGFEAADAKT 95
G+ T
Sbjct 1267 LGYMVRKGAT 1276
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 28/54 (51%), Gaps = 2/54 (3%)
Query 40 CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADA 93
C G+ P G AC+DVDEC A C CVN+ GSF C C G +D+
Sbjct 232 CRRGFIPNIHTG-ACQDVDECQAVPGLCQ-GGSCVNMVGSFHCRCPVGHRLSDS 283
Score = 37.0 bits (84), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 30/65 (46%), Gaps = 2/65 (3%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N G Y C+C G+ S C D +ECA C SA C N G F C C +GF
Sbjct 2542 CQNQLGGYRCSCPQGFT-QHSQWAQCVDENECALSPPTCG-SASCRNTLGGFRCVCPSGF 2599
Query 89 EAADA 93
+ A
Sbjct 2600 DFDQA 2604
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 21/42 (50%), Gaps = 1/42 (2%)
Query 47 ASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
A G C DV+EC + C + +CVN GSF C C G
Sbjct 860 ARMTGVTCDDVNECESFPGVC-PNGRCVNTAGSFRCECPEGL 900
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 28/61 (45%), Gaps = 2/61 (3%)
Query 32 TEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAA 91
TE +C G++P +D+DEC C CVN GSF+C C G+ +
Sbjct 1540 TEYRTLCPGGEGFQPNRIT-VILEDIDECQELPGLCQ-GGDCVNTFGSFQCECPPGYHLS 1597
Query 92 D 92
+
Sbjct 1598 E 1598
Score = 27.7 bits (60), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 10/31 (32%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 19 AHSSTSLHASCANTEGSYVCTCNPGYEPASS 49
A ++ C NT GS+ C C+ G++P+ +
Sbjct 2258 AQPDLCVNGRCVNTAGSFRCDCDEGFQPSPT 2288
> hsa:976 CD97, TM7LN1; CD97 molecule; K08446 CD97 molecule
Length=786
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 34/79 (43%), Positives = 41/79 (51%), Gaps = 9/79 (11%)
Query 27 ASCANTEGSYVCTCNPGYEPAS-------SDGHACKDVDECAAGTAECHVSAQCVNVDGS 79
+ C NTEGSY C C+PGYEP S + C+DVDEC C CVN GS
Sbjct 80 SDCWNTEGSYDCVCSPGYEPVSGAKTFKNESENTCQDVDECQQNPRLCKSYGTCVNTLGS 139
Query 80 FECTCMAGFE--AADAKTC 96
+ C C+ GF+ D K C
Sbjct 140 YTCQCLPGFKFIPEDPKVC 158
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 37/64 (57%), Gaps = 0/64 (0%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ +C NT GSY C C PG++ D C DVDEC++G +C S C N GS+ C C
Sbjct 130 YGTCVNTLGSYTCQCLPGFKFIPEDPKVCTDVDECSSGQHQCDSSTVCFNTVGSYSCRCR 189
Query 86 AGFE 89
G++
Sbjct 190 PGWK 193
Score = 31.2 bits (69), Expect = 0.89, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 16/28 (57%), Gaps = 0/28 (0%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKD 56
C NT GSY C C PG++P + KD
Sbjct 177 CFNTVGSYSCRCRPGWKPRHGIPNNQKD 204
> mmu:268977 Ltbp1, 9430031G15Rik, 9830146M04, Ltbp-1, Tgfb; latent
transforming growth factor beta binding protein 1; K08023
latent transforming growth factor beta binding protein
Length=1712
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 33/69 (47%), Positives = 44/69 (63%), Gaps = 3/69 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ SC N EGSY+C+C+ GY P H C+D+DEC G C ++ QC N DGSF CTC
Sbjct 1042 NGSCTNLEGSYMCSCHRGYSPTPDHRH-CQDIDECQQGNL-C-MNGQCRNTDGSFRCTCG 1098
Query 86 AGFEAADAK 94
G++ + AK
Sbjct 1099 QGYQLSAAK 1107
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 43/72 (59%), Gaps = 2/72 (2%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H C NTEGS+ C CN GY AS G C+D++EC ++ C C+N GS++CTC
Sbjct 1124 HGQCRNTEGSFQCVCNQGYR-ASVLGDHCEDINECLEDSSVCQ-GGDCINTAGSYDCTCP 1181
Query 86 AGFEAADAKTCK 97
GF+ D K C+
Sbjct 1182 DGFQLNDNKGCQ 1193
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/72 (43%), Positives = 39/72 (54%), Gaps = 3/72 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H C NT+GS+ C C G+ S+DG C+D+DEC T C C N GSF C C
Sbjct 1207 HGECLNTQGSFHCVCEQGFS-ISADGRTCEDIDECVNNTV-CDSHGFCDNTAGSFRCLCY 1264
Query 86 AGFEAA-DAKTC 96
GF+A D + C
Sbjct 1265 QGFQAPQDGQGC 1276
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 32/78 (41%), Positives = 43/78 (55%), Gaps = 4/78 (5%)
Query 21 SSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSF 80
SS C NT GSY CTC G++ +D C+D++ECA C +C+N GSF
Sbjct 1161 SSVCQGGDCINTAGSYDCTCPDGFQ--LNDNKGCQDINECAQ-PGLCGSHGECLNTQGSF 1217
Query 81 ECTCMAGFE-AADAKTCK 97
C C GF +AD +TC+
Sbjct 1218 HCVCEQGFSISADGRTCE 1235
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 39/67 (58%), Gaps = 3/67 (4%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
++ C NT+GS+ CTC GY+ S+ C+D+DEC H QC N +GSF+C C
Sbjct 1082 MNGQCRNTDGSFRCTCGQGYQ-LSAAKDQCEDIDECEHHHLCSH--GQCRNTEGSFQCVC 1138
Query 85 MAGFEAA 91
G+ A+
Sbjct 1139 NQGYRAS 1145
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 27/59 (45%), Positives = 33/59 (55%), Gaps = 2/59 (3%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
H C NT GS+ C C G++ A DG C DV+EC + C A C NV+GSF C C
Sbjct 1249 HGFCDNTAGSFRCLCYQGFQ-APQDGQGCVDVNECELLSGVCG-EAFCENVEGSFLCVC 1305
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 33/67 (49%), Gaps = 2/67 (2%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N Y C C GY+ S C D+DECA C +C N +GSF C C AGF
Sbjct 881 CINLPVRYTCICYEGYK-FSEQLRKCVDIDECAQVRHLCS-QGRCENTEGSFLCVCPAGF 938
Query 89 EAADAKT 95
A++ T
Sbjct 939 MASEEGT 945
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 38/73 (52%), Gaps = 4/73 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT-C 84
C NTEGS++C C G+ AS +G C DVDEC + +C+N G+F C C
Sbjct 920 QGRCENTEGSFLCVCPAGFM-ASEEGTNCIDVDECL--RPDMCRDGRCINTAGAFRCEYC 976
Query 85 MAGFEAADAKTCK 97
+G+ + C+
Sbjct 977 DSGYRMSRRGYCE 989
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 34/69 (49%), Gaps = 3/69 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHV--SAQCVNVDGSFECT 83
+ C + Y C C GY C DV+EC+ + +A+C+N +GS++C
Sbjct 1624 NGRCVRVQEGYTCDCFDGYH-LDMAKMTCVDVNECSELNNRMSLCKNAKCINTEGSYKCL 1682
Query 84 CMAGFEAAD 92
C+ G+ +D
Sbjct 1683 CLPGYIPSD 1691
Score = 35.4 bits (80), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 23/58 (39%), Positives = 30/58 (51%), Gaps = 7/58 (12%)
Query 29 CANTEGSYVCTCNPG--YEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
C NT+ Y C C G Y+P C D+DEC + C + QCVN +GS+ C C
Sbjct 1432 CLNTQPGYECYCKQGTYYDPVKLQ---CFDMDECQDPNS-C-IDGQCVNTEGSYNCFC 1484
Score = 35.4 bits (80), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 28/54 (51%), Gaps = 4/54 (7%)
Query 1 LPMVFRQCVFVPQGEASAAHSSTSL--HASCANTEGSYVCTCNPGYEPASSDGH 52
L M CV V E S ++ SL +A C NTEGSY C C PGY P+ +
Sbjct 1644 LDMAKMTCVDV--NECSELNNRMSLCKNAKCINTEGSYKCLCLPGYIPSDKPNY 1695
Score = 27.7 bits (60), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 21/35 (60%), Gaps = 2/35 (5%)
Query 54 CKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C+D++EC C + +C+N GS+ C+C GF
Sbjct 616 CQDINECQL-QGVC-PNGECLNTMGSYRCSCKMGF 648
> hsa:4052 LTBP1, MGC163161; latent transforming growth factor
beta binding protein 1; K08023 latent transforming growth factor
beta binding protein
Length=1395
Score = 64.3 bits (155), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 33/78 (42%), Positives = 44/78 (56%), Gaps = 2/78 (2%)
Query 20 HSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGS 79
H H C NTEGS+ C C+ GY AS G C+D++EC + C C+N GS
Sbjct 800 HRHLCAHGQCRNTEGSFQCVCDQGYR-ASGLGDHCEDINECLEDKSVCQ-RGDCINTAGS 857
Query 80 FECTCMAGFEAADAKTCK 97
++CTC GF+ D KTC+
Sbjct 858 YDCTCPDGFQLDDNKTCQ 875
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/69 (43%), Positives = 43/69 (62%), Gaps = 3/69 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C+N EGSY+C+C+ GY + D C+D+DEC G V+ QC N +GSF CTC
Sbjct 724 NGDCSNLEGSYMCSCHKGYT-RTPDHKHCRDIDECQQGNL--CVNGQCKNTEGSFRCTCG 780
Query 86 AGFEAADAK 94
G++ + AK
Sbjct 781 QGYQLSAAK 789
Score = 54.7 bits (130), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 31/72 (43%), Positives = 38/72 (52%), Gaps = 3/72 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
C NTEGS+ C C G+ S+DG C+D+DEC T C C N GSF C C
Sbjct 889 QGECLNTEGSFHCVCQQGFS-ISADGRTCEDIDECVNNTV-CDSHGFCDNTAGSFRCLCY 946
Query 86 AGFEAA-DAKTC 96
GF+A D + C
Sbjct 947 QGFQAPQDGQGC 958
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 40/73 (54%), Gaps = 4/73 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
C NT GSY CTC G++ D C+D++EC C +C+N +GSF C C
Sbjct 848 RGDCINTAGSYDCTCPDGFQ--LDDNKTCQDINECEH-PGLCGPQGECLNTEGSFHCVCQ 904
Query 86 AGFE-AADAKTCK 97
GF +AD +TC+
Sbjct 905 QGFSISADGRTCE 917
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 28/67 (41%), Positives = 39/67 (58%), Gaps = 3/67 (4%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
++ C NTEGS+ CTC GY+ S+ C+D+DEC H QC N +GSF+C C
Sbjct 764 VNGQCKNTEGSFRCTCGQGYQ-LSAAKDQCEDIDECQHRHLCAH--GQCRNTEGSFQCVC 820
Query 85 MAGFEAA 91
G+ A+
Sbjct 821 DQGYRAS 827
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 27/59 (45%), Positives = 33/59 (55%), Gaps = 2/59 (3%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
H C NT GS+ C C G++ A DG C DV+EC + C A C NV+GSF C C
Sbjct 931 HGFCDNTAGSFRCLCYQGFQ-APQDGQGCVDVNECELLSGVCG-EAFCENVEGSFLCVC 987
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/67 (38%), Positives = 31/67 (46%), Gaps = 2/67 (2%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N Y C C GY S C D+DEC C +C N +GSF C C AGF
Sbjct 563 CINLPVRYTCICYEGYR-FSEQQRKCVDIDECTQVQHLCS-QGRCENTEGSFLCICPAGF 620
Query 89 EAADAKT 95
A++ T
Sbjct 621 MASEEGT 627
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 36/73 (49%), Gaps = 4/73 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT-C 84
C NTEGS++C C G+ AS +G C DVDEC + CVN G+F C C
Sbjct 602 QGRCENTEGSFLCICPAGFM-ASEEGTNCIDVDECLR--PDVCGEGHCVNTVGAFRCEYC 658
Query 85 MAGFEAADAKTCK 97
+G+ C+
Sbjct 659 DSGYRMTQRGRCE 671
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 34/70 (48%), Gaps = 3/70 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHV--SAQCVNVDGSFECT 83
+ C + Y C C GY + C DV+EC + +A+C+N DGS++C
Sbjct 1307 NGRCVRVQEGYTCDCFDGYH-LDTAKMTCVDVNECDELNNRMSLCKNAKCINTDGSYKCL 1365
Query 84 CMAGFEAADA 93
C+ G+ +D
Sbjct 1366 CLPGYVPSDK 1375
Score = 35.4 bits (80), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 28/92 (30%), Positives = 39/92 (42%), Gaps = 22/92 (23%)
Query 10 FVPQGEASAAHSSTSL---------------HASCANTEGSYVCTCNPG--YEPASSDGH 52
FVP GE+S+ + + C NT Y C C G Y+P
Sbjct 1081 FVPAGESSSEAGGENYKDADECLLFGQEICKNGFCLNTRPGYECYCKQGTYYDPVKL--- 1137
Query 53 ACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
C D+DEC ++ + QCVN +GS+ C C
Sbjct 1138 QCFDMDECQDPSS--CIDGQCVNTEGSYNCFC 1167
Score = 33.1 bits (74), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 23/45 (51%), Gaps = 0/45 (0%)
Query 8 CVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGH 52
CV V + + S +A C NT+GSY C C PGY P+ +
Sbjct 1334 CVDVNECDELNNRMSLCKNAKCINTDGSYKCLCLPGYVPSDKPNY 1378
Score = 31.6 bits (70), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 28/49 (57%), Gaps = 4/49 (8%)
Query 40 CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C PGY+ ++ C+D++EC C + +C+N GS+ CTC GF
Sbjct 286 CLPGYKRVNNT--FCQDINECQL-QGVC-PNGECLNTMGSYRCTCKIGF 330
> hsa:2201 FBN2, CCA, DA9; fibrillin 2
Length=2912
Score = 64.3 bits (155), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 33/72 (45%), Positives = 44/72 (61%), Gaps = 4/72 (5%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
+HASC N GS+ C+C G+ +G C D+DEC+ GT +C ++AQCVN GS+ C C
Sbjct 1380 MHASCLNIPGSFKCSCREGW---IGNGIKCIDLDECSNGTHQCSINAQCVNTPGSYRCAC 1436
Query 85 MAGFEAADAKTC 96
GF D TC
Sbjct 1437 SEGF-TGDGFTC 1447
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 35/84 (41%), Positives = 49/84 (58%), Gaps = 5/84 (5%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E S S++A C NT GSY C C+ G+ + DG C DVDECA C + QC+
Sbjct 1411 ECSNGTHQCSINAQCVNTPGSYRCACSEGF---TGDGFTCSDVDECAENINLCE-NGQCL 1466
Query 75 NVDGSFECTCMAGF-EAADAKTCK 97
NV G++ C C GF A+D+++C+
Sbjct 1467 NVPGAYRCECEMGFTPASDSRSCQ 1490
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 33/77 (42%), Positives = 43/77 (55%), Gaps = 3/77 (3%)
Query 21 SSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSF 80
S+ + C NT GSY C C+ G++ + AC D+DEC C + +CVN DGSF
Sbjct 541 SNPCTNGDCVNTPGSYYCKCHAGFQRTPTK-QACIDIDECIQNGVLCK-NGRCVNTDGSF 598
Query 81 ECTCMAGFE-AADAKTC 96
+C C AGFE D K C
Sbjct 599 QCICNAGFELTTDGKNC 615
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 47/92 (51%), Gaps = 4/92 (4%)
Query 3 MVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAA 62
M + C+ V + + +S+ + C NT+GS++C C GY C DVDEC
Sbjct 1318 MDMKTCIDVNECDL---NSNICMFGECENTKGSFICHCQLGYS-VKKGTTGCTDVDECEI 1373
Query 63 GTAECHVSAQCVNVDGSFECTCMAGFEAADAK 94
G C + A C+N+ GSF+C+C G+ K
Sbjct 1374 GAHNCDMHASCLNIPGSFKCSCREGWIGNGIK 1405
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 41/72 (56%), Gaps = 4/72 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N G+Y C+CNPGY+ A+ D C D+DEC C QC N +GS+EC+C
Sbjct 1213 NGKCVNMIGTYQCSCNPGYQ-ATPDRQGCTDIDECMIMNGGC--DTQCTNSEGSYECSCS 1269
Query 86 AGFE-AADAKTC 96
G+ D ++C
Sbjct 1270 EGYALMPDGRSC 1281
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 31/71 (43%), Positives = 39/71 (54%), Gaps = 3/71 (4%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C N+EGSY C+C+ GY DG +C D+DEC C QC N+ G + C C
Sbjct 1255 TQCTNSEGSYECSCSEGYA-LMPDGRSCADIDECENNPDICD-GGQCTNIPGEYRCLCYD 1312
Query 87 GFEAA-DAKTC 96
GF A+ D KTC
Sbjct 1313 GFMASMDMKTC 1323
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 39/66 (59%), Gaps = 3/66 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT+GS+ C CN G+E ++DG C D DEC T ++ C+N DGSF+C C
Sbjct 588 NGRCVNTDGSFQCICNAGFE-LTTDGKNCVDHDECT--TTNMCLNGMCINEDGSFKCICK 644
Query 86 AGFEAA 91
GF A
Sbjct 645 PGFVLA 650
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/63 (44%), Positives = 34/63 (53%), Gaps = 2/63 (3%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N GSY C CN GYEP +S G C D+DEC C + C N GS+ CTC
Sbjct 781 NGICENLRGSYRCNCNSGYEPDAS-GRNCIDIDECLVNRLLCD-NGLCRNTPGSYSCTCP 838
Query 86 AGF 88
G+
Sbjct 839 PGY 841
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 41/72 (56%), Gaps = 3/72 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ +C NT GSY C C PG+E ++ C D+DEC++ + + +C N GSF+C C
Sbjct 1945 NGTCKNTVGSYNCLCYPGFELTHNN--DCLDIDECSSFFGQVCRNGRCFNEIGSFKCLCN 2002
Query 86 AGFE-AADAKTC 96
G+E D K C
Sbjct 2003 EGYELTPDGKNC 2014
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 34/70 (48%), Gaps = 3/70 (4%)
Query 19 AHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDG 78
+ S + C NTEGSY C+C GY DG CKD+DEC C CVN G
Sbjct 2496 SQSPKPCNYICKNTEGSYQCSCPRGYV-LQEDGKTCKDLDECQTKQHNCQF--LCVNTLG 2552
Query 79 SFECTCMAGF 88
F C C GF
Sbjct 2553 GFTCKCPPGF 2562
Score = 50.8 bits (120), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 41/73 (56%), Gaps = 5/73 (6%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ +C N GS+ C CN G+EP C+D++ECA C + +C+N GS+ECTC
Sbjct 2225 NGTCTNVIGSFECNCNEGFEPGPM--MNCEDINECAQNPLLC--AFRCMNTFGSYECTCP 2280
Query 86 AGFEAA-DAKTCK 97
G+ D K CK
Sbjct 2281 IGYALREDQKMCK 2293
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/76 (38%), Positives = 37/76 (48%), Gaps = 2/76 (2%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E S+ + C N GS+ C CN GYE + DG C D +EC A C C
Sbjct 1976 ECSSFFGQVCRNGRCFNEIGSFKCLCNEGYE-LTPDGKNCIDTNECVALPGSCS-PGTCQ 2033
Query 75 NVDGSFECTCMAGFEA 90
N++GSF C C G+E
Sbjct 2034 NLEGSFRCICPPGYEV 2049
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 39/80 (48%), Gaps = 2/80 (2%)
Query 16 ASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVN 75
A H S C N G+++C C PG DG C D +EC C + +CVN
Sbjct 2299 AEGLHDCESRGMMCKNLIGTFMCICPPGMA-RRPDGEGCVDENECRTKPGICE-NGRCVN 2356
Query 76 VDGSFECTCMAGFEAADAKT 95
+ GS+ C C GF+++ + T
Sbjct 2357 IIGSYRCECNEGFQSSSSGT 2376
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 31/94 (32%), Positives = 42/94 (44%), Gaps = 4/94 (4%)
Query 1 LPMVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDEC 60
L M R C + + S + + C NT GS+ C C GYE C D+DEC
Sbjct 1106 LDMEERNCTDIDECRISPDLCGSGI---CVNTPGSFECECFEGYESGFMMMKNCMDIDEC 1162
Query 61 AAGTAECHVSAQCVNVDGSFECTCMAGFEAADAK 94
C CVN +GSF+C C G E + ++
Sbjct 1163 ERNPLLCR-GGTCVNTEGSFQCDCPLGHELSPSR 1195
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 38/73 (52%), Gaps = 4/73 (5%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
L+ C N +GS+ C C PG+ A +G C DVDEC T ++ C+N +GSF C C
Sbjct 628 LNGMCINEDGSFKCICKPGFVLA-PNGRYCTDVDECQ--TPGICMNGHCINSEGSFRCDC 684
Query 85 MAGFEAA-DAKTC 96
G D + C
Sbjct 685 PPGLAVGMDGRVC 697
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 35/72 (48%), Gaps = 3/72 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT+GS+ C C GY G C D DEC+ G + C NV GSFEC C
Sbjct 2184 NGQCINTDGSFRCECPMGYN-LDYTGVRCVDTDECSIGNP--CGNGTCTNVIGSFECNCN 2240
Query 86 AGFEAADAKTCK 97
GFE C+
Sbjct 2241 EGFEPGPMMNCE 2252
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 34/66 (51%), Gaps = 2/66 (3%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT GS+ C CN G+ + C D+DEC C S CVN GSFEC C
Sbjct 1086 YGKCRNTIGSFKCRCNSGFA-LDMEERNCTDIDECRISPDLCG-SGICVNTPGSFECECF 1143
Query 86 AGFEAA 91
G+E+
Sbjct 1144 EGYESG 1149
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/61 (45%), Positives = 32/61 (52%), Gaps = 2/61 (3%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSA-QCVNVDGSFECTCMAG 87
C NT GSY CTC GY D CKD+DECA G +C C N+ G+F C C G
Sbjct 2268 CMNTFGSYECTCPIGYA-LREDQKMCKDLDECAEGLHDCESRGMMCKNLIGTFMCICPPG 2326
Query 88 F 88
Sbjct 2327 M 2327
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/92 (29%), Positives = 50/92 (54%), Gaps = 5/92 (5%)
Query 3 MVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAA 62
M+ + C+ + + E + +C NTEGS+ C C G+E + S C D++EC+
Sbjct 1151 MMMKNCMDIDECERNPL---LCRGGTCVNTEGSFQCDCPLGHELSPSR-EDCVDINECSL 1206
Query 63 GTAECHVSAQCVNVDGSFECTCMAGFEAADAK 94
C + +CVN+ G+++C+C G++A +
Sbjct 1207 SDNLCR-NGKCVNMIGTYQCSCNPGYQATPDR 1237
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 40/73 (54%), Gaps = 4/73 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT GS+ C C GY S G +C D+DEC+ C+ C N +GS++C+C
Sbjct 2462 NGQCINTMGSFRCFCKVGYTTDIS-GTSCIDLDECSQSPKPCNYI--CKNTEGSYQCSCP 2518
Query 86 AGFEAA-DAKTCK 97
G+ D KTCK
Sbjct 2519 RGYVLQEDGKTCK 2531
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 37/71 (52%), Gaps = 1/71 (1%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N GS+ C C G+ + C+D+DEC+ G C +A C+N GS+ C C
Sbjct 1821 NGVCINQIGSFRCECPTGFS-YNDLLLVCEDIDECSNGDNLCQRNADCINSPGSYRCECA 1879
Query 86 AGFEAADAKTC 96
AGF+ + C
Sbjct 1880 AGFKLSPNGAC 1890
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 34/66 (51%), Gaps = 4/66 (6%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
+C N EGS+ C C PGYE S + C D++EC C + C N G F+C C
Sbjct 2030 GTCQNLEGSFRCICPPGYEVKSEN---CIDINECDEDPNIC-LFGSCTNTPGGFQCLCPP 2085
Query 87 GFEAAD 92
GF +D
Sbjct 2086 GFVLSD 2091
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 30/76 (39%), Positives = 39/76 (51%), Gaps = 7/76 (9%)
Query 20 HSSTSLHASCANTEGSYVCTCNPGY-EPASSDGHACKDVDECAAGTAECHVSAQCVNVDG 78
H++ L+ C T SY C CN GY + A+ D C DVDEC + + CVN G
Sbjct 501 HANLCLNGRCIPTVSSYRCECNMGYKQDANGD---CIDVDECTSNPC---TNGDCVNTPG 554
Query 79 SFECTCMAGFEAADAK 94
S+ C C AGF+ K
Sbjct 555 SYYCKCHAGFQRTPTK 570
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 34/64 (53%), Gaps = 3/64 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N G+Y C C G+ PA SD +C+D+DEC+ V C N+ G F C C
Sbjct 1462 NGQCLNVPGAYRCECEMGFTPA-SDSRSCQDIDECSFQNI--CVFGTCNNLPGMFHCICD 1518
Query 86 AGFE 89
G+E
Sbjct 1519 DGYE 1522
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 27/71 (38%), Positives = 34/71 (47%), Gaps = 4/71 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H C + +GSY C C+ G++ AS D C DVDEC + C N GS+ C C
Sbjct 1905 HGLCVDLQGSYQCICHNGFK-ASQDQTMCMDVDECERHPCG---NGTCKNTVGSYNCLCY 1960
Query 86 AGFEAADAKTC 96
GFE C
Sbjct 1961 PGFELTHNNDC 1971
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 35/65 (53%), Gaps = 3/65 (4%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
+ +C N G + C C+ GYE + G+ C D+DECA C V+ CVN G +EC C
Sbjct 1502 VFGTCNNLPGMFHCICDDGYELDRTGGN-CTDIDECAD-PINC-VNGLCVNTPGRYECNC 1558
Query 85 MAGFE 89
F+
Sbjct 1559 PPDFQ 1563
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 43/81 (53%), Gaps = 3/81 (3%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E S + +A C N+ GSY C C G++ S +G AC D +EC C CV
Sbjct 1853 ECSNGDNLCQRNADCINSPGSYRCECAAGFK-LSPNG-ACVDRNECLEIPNVCS-HGLCV 1909
Query 75 NVDGSFECTCMAGFEAADAKT 95
++ GS++C C GF+A+ +T
Sbjct 1910 DLQGSYQCICHNGFKASQDQT 1930
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 29/60 (48%), Gaps = 3/60 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT G + C C PG+ + AC D +EC + + C C N GSF C C GF
Sbjct 2547 CVNTLGGFTCKCPPGFTQHHT---ACIDNNECGSQPSLCGAKGICQNTPGSFSCECQRGF 2603
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 31/69 (44%), Gaps = 2/69 (2%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C N G Y C C G+ AS D C DV+EC + C + +C N GSF C C
Sbjct 1297 GQCTNIPGEYRCLCYDGFM-ASMDMKTCIDVNECDLNSNIC-MFGECENTKGSFICHCQL 1354
Query 87 GFEAADAKT 95
G+ T
Sbjct 1355 GYSVKKGTT 1363
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 29/54 (53%), Gaps = 2/54 (3%)
Query 40 CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADA 93
C G+ P G AC+DVDEC A C C+N GSFEC C AG + ++
Sbjct 261 CRRGFIPNIRTG-ACQDVDECQAIPGICQ-GGNCINTVGSFECRCPAGHKQSET 312
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 40/84 (47%), Gaps = 7/84 (8%)
Query 5 FRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGT 64
+ QCV E ++ + ASC NT GSY C C G+ AC DV+EC++
Sbjct 2649 WNQCV----DENECSNPNACGSASCYNTLGSYKCACPSGFS-FDQFSSACHDVNECSSSK 2703
Query 65 AECHVSAQCVNVDGSFECTCMAGF 88
C+ C N +G + C C G+
Sbjct 2704 NPCNYG--CSNTEGGYLCGCPPGY 2725
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 33/62 (53%), Gaps = 2/62 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
+C NT GS+ C C G++ S C+D+DEC+ C + +C N GS+ C C
Sbjct 290 GNCINTVGSFECRCPAGHK-QSETTQKCEDIDECSIIPGICE-TGECSNTVGSYFCVCPR 347
Query 87 GF 88
G+
Sbjct 348 GY 349
Score = 37.4 bits (85), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 23/58 (39%), Positives = 29/58 (50%), Gaps = 2/58 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
+C NT GS+ C C GY S D C+D+DEC A C C N G++ C C
Sbjct 1664 GNCINTFGSFQCECPQGYY-LSEDTRICEDIDECFAHPGVCG-PGTCYNTLGNYTCIC 1719
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 25/78 (32%), Positives = 34/78 (43%), Gaps = 4/78 (5%)
Query 11 VPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVS 70
+ E + S C NT GS+ C C G+ + G C+DVDEC G C
Sbjct 2570 IDNNECGSQPSLCGAKGICQNTPGSFSCECQRGFS-LDATGLNCEDVDEC-DGNHRCQHG 2627
Query 71 AQCVNVDGSFECTCMAGF 88
Q N+ G + C C G+
Sbjct 2628 CQ--NILGGYRCGCPQGY 2643
Score = 34.7 bits (78), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 28/52 (53%), Gaps = 7/52 (13%)
Query 37 VCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
+C PGY ++DG +D+DEC C + QC+N GSF C C G+
Sbjct 2436 ICPHGPGY---TTDG---RDIDECKVMPNLC-TNGQCINTMGSFRCFCKVGY 2480
Score = 32.3 bits (72), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 29/60 (48%), Gaps = 3/60 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N G Y C C GY + C D +EC+ A SA C N GS++C C +GF
Sbjct 2628 CQNILGGYRCGCPQGYI-QHYQWNQCVDENECSNPNA--CGSASCYNTLGSYKCACPSGF 2684
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 21/42 (50%), Gaps = 1/42 (2%)
Query 47 ASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
A G C+DV+EC C + +CVN GSF C C G
Sbjct 946 ARIKGVTCEDVNECEVFPGVCP-NGRCVNSKGSFHCECPEGL 986
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 23/44 (52%), Gaps = 2/44 (4%)
Query 55 KDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAA-DAKTCK 97
+D+DEC C C+N GSF+C C G+ + D + C+
Sbjct 1649 EDIDECQELPGLCQ-GGNCINTFGSFQCECPQGYYLSEDTRICE 1691
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 19/39 (48%), Gaps = 1/39 (2%)
Query 19 AHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDV 57
AH +C NT G+Y C C P Y + GH C D+
Sbjct 1698 AHPGVCGPGTCYNTLGNYTCICPPEYMQVNG-GHNCMDM 1735
Score = 28.5 bits (62), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 18/35 (51%), Gaps = 1/35 (2%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDE 59
L SC NT G + C C PG+ S +G C D +
Sbjct 2068 LFGSCTNTPGGFQCLCPPGFV-LSDNGRRCFDTRQ 2101
Score = 28.1 bits (61), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 17/30 (56%), Gaps = 1/30 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKD 56
C+NT GSY C C GY S+DG C D
Sbjct 332 GECSNTVGSYFCVCPRGYV-TSTDGSRCID 360
> mmu:26364 Cd97, AA409984, TM7LN1; CD97 antigen; K08446 CD97
molecule
Length=724
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 30/70 (42%), Positives = 41/70 (58%), Gaps = 7/70 (10%)
Query 27 ASCANTEGSYVCTCNPGYEPAS-------SDGHACKDVDECAAGTAECHVSAQCVNVDGS 79
A C N+EGSY C CN GY+ S + C+DVDEC++G +CH S C N GS
Sbjct 84 AMCKNSEGSYTCVCNLGYKLLSGAESFVNESENTCQDVDECSSGQHQCHNSTVCKNTVGS 143
Query 80 FECTCMAGFE 89
++C C G++
Sbjct 144 YKCHCRPGWK 153
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 18/56 (32%), Positives = 29/56 (51%), Gaps = 4/56 (7%)
Query 38 CTCNPGY----EPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFE 89
C C PG+ E ++ +C+D++EC C A C N +GS+ C C G++
Sbjct 47 CVCKPGFSSEKELITNPAESCEDINECLLPGFSCGDFAMCKNSEGSYTCVCNLGYK 102
Score = 32.7 bits (73), Expect = 0.30, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 17/34 (50%), Gaps = 0/34 (0%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPAS 48
E S+ C NT GSY C C PG++P S
Sbjct 123 ECSSGQHQCHNSTVCKNTVGSYKCHCRPGWKPTS 156
> dre:571786 fbn2b, fbn2; fibrillin 2b
Length=2868
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 38/95 (40%), Positives = 52/95 (54%), Gaps = 3/95 (3%)
Query 3 MVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAA 62
M +RQ V + S+ ++ C NT+GSY C C+ GY+ + AC D+DEC
Sbjct 483 MGYRQDVRGECIDVDECVSNPCVNGDCVNTQGSYHCKCHEGYQ-GTPTKQACIDIDECIV 541
Query 63 GTAECHVSAQCVNVDGSFECTCMAGFEAA-DAKTC 96
C + +CVN DGSF+C C AGFE + D K C
Sbjct 542 NGVMCR-NGRCVNTDGSFQCICNAGFEISPDGKNC 575
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 35/92 (38%), Positives = 46/92 (50%), Gaps = 4/92 (4%)
Query 3 MVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAA 62
M R C+ V + + + + LH C NT+GS++C C GY C DVDEC
Sbjct 1278 MDMRTCIDVNECDLNP---NICLHGDCENTKGSFICHCQLGYF-VKKGSTGCTDVDECEI 1333
Query 63 GTAECHVSAQCVNVDGSFECTCMAGFEAADAK 94
G C + A CVN GSF+C C G+E K
Sbjct 1334 GAHNCDLHAACVNAPGSFKCRCRDGWEGDGIK 1365
Score = 58.9 bits (141), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 38/66 (57%), Gaps = 3/66 (4%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
LHA+C N GS+ C C G+E DG C DVDEC C+ +A+C+N GS+ C+C
Sbjct 1340 LHAACVNAPGSFKCRCRDGWE---GDGIKCIDVDECVTEEHNCNPNAECLNTPGSYRCSC 1396
Query 85 MAGFEA 90
GF
Sbjct 1397 KEGFNG 1402
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 39/66 (59%), Gaps = 3/66 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT+GS+ C CN G+E S DG C D DECA T ++ C+N DGSF+C C
Sbjct 548 NGRCVNTDGSFQCICNAGFE-ISPDGKNCIDHDECA--TTNMCLNGMCINEDGSFKCICK 604
Query 86 AGFEAA 91
GF A
Sbjct 605 PGFALA 610
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 42/72 (58%), Gaps = 3/72 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ +C NT GSY C C PG+E ++ C D+DEC+ ++ + QC+N GSF+C C+
Sbjct 1905 NGTCKNTVGSYNCLCFPGFELTHNN--DCMDIDECSVHASQVCRNGQCINNMGSFKCLCL 1962
Query 86 AGFEAA-DAKTC 96
G+ D K C
Sbjct 1963 DGYNLTPDGKNC 1974
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 41/73 (56%), Gaps = 5/73 (6%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+A C NT GSY C+C G+ + DG +C D+DECA C + QC+N G + C C
Sbjct 1382 NAECLNTPGSYRCSCKEGF---NGDGFSCSDMDECADNVNLCE-NGQCLNAPGGYRCECE 1437
Query 86 AGFEAA-DAKTCK 97
GF D+K C+
Sbjct 1438 MGFTPTEDSKACQ 1450
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/92 (32%), Positives = 50/92 (54%), Gaps = 5/92 (5%)
Query 3 MVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAA 62
M+ + C+ + + E +C NTEGSY C C PG++ S++ AC+DV+EC
Sbjct 1111 MMMKNCMDIDECERDPL---LCRGGTCLNTEGSYECDCPPGHQ-LSTEASACEDVNECQL 1166
Query 63 GTAECHVSAQCVNVDGSFECTCMAGFEAADAK 94
C QCVN+ G+++C+C G++ +
Sbjct 1167 SDNLCK-HGQCVNMVGTYQCSCETGYQVTPDR 1197
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 37/70 (52%), Gaps = 3/70 (4%)
Query 19 AHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDG 78
+ S + C NTEGSY+C+C GY DG CKD+DEC+ C CVN G
Sbjct 2456 SQSPKPCNFLCKNTEGSYLCSCPRGY-ILQPDGKTCKDLDECSTKQHNCQF--LCVNTIG 2512
Query 79 SFECTCMAGF 88
F C C +GF
Sbjct 2513 GFTCKCPSGF 2522
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 37/80 (46%), Gaps = 2/80 (2%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E S S + C N GS+ C C GY + DG C D++EC C C
Sbjct 1936 ECSVHASQVCRNGQCINNMGSFKCLCLDGYN-LTPDGKNCVDINECVTLPGACQ-PGTCQ 1993
Query 75 NVDGSFECTCMAGFEAADAK 94
N+DGSF C C G+E + K
Sbjct 1994 NLDGSFRCICPPGYEVQNDK 2013
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 43/79 (54%), Gaps = 4/79 (5%)
Query 19 AHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDG 78
A ++ L+ C N +GS+ C C PG+ A +G C D+DEC T+ ++ +CVN +G
Sbjct 582 ATTNMCLNGMCINEDGSFKCICKPGFALA-PNGRYCTDIDECH--TSGICMNGRCVNTEG 638
Query 79 SFECTCMAGFEA-ADAKTC 96
SF C C G D + C
Sbjct 639 SFRCECPPGLAIDVDGRVC 657
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 40/73 (54%), Gaps = 3/73 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N GSY C CN GYE +S G C D++EC C + C N GS+ C+C
Sbjct 741 NGICENLRGSYRCICNIGYESDAS-GKNCVDINECLVNRLLCD-NGMCRNTPGSYTCSCP 798
Query 86 AGFE-AADAKTCK 97
GF AD++TC+
Sbjct 799 KGFSFKADSETCE 811
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/72 (37%), Positives = 37/72 (51%), Gaps = 4/72 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H C N G+Y C+C GY+ + D C D+DEC C C N +GS+EC+C
Sbjct 1173 HGQCVNMVGTYQCSCETGYQ-VTPDRQGCVDIDECTIMNGGC--DTHCTNSEGSYECSCS 1229
Query 86 AGFEAA-DAKTC 96
G+ D +TC
Sbjct 1230 EGYALMPDLRTC 1241
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 40/71 (56%), Gaps = 1/71 (1%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N GS+ C C G+ ++ C+D+DEC +G C +A C+N+ GS+ C C
Sbjct 1781 NGVCINQIGSFRCECPMGFS-YNNILLICEDIDECNSGDNLCQRNANCINIPGSYRCECS 1839
Query 86 AGFEAADAKTC 96
AGF+ + + C
Sbjct 1840 AGFKLSPSGAC 1850
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 29/69 (42%), Positives = 37/69 (53%), Gaps = 3/69 (4%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N+EGSY C+C+ GY D C D+DEC C QC N+ G + C C GF
Sbjct 1217 CTNSEGSYECSCSEGYA-LMPDLRTCADIDECEDTPDICD-GGQCTNIPGEYRCLCYDGF 1274
Query 89 EAA-DAKTC 96
A+ D +TC
Sbjct 1275 MASMDMRTC 1283
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 39/80 (48%), Gaps = 2/80 (2%)
Query 16 ASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVN 75
A S +C N G+++C C PG DG C D++EC + C + +CVN
Sbjct 2259 AEGLDDCASRGMACKNQIGTFMCICPPGMT-RRPDGEGCMDLNECRSKPGICK-NGRCVN 2316
Query 76 VDGSFECTCMAGFEAADAKT 95
GS+ C C GFEA+ T
Sbjct 2317 TVGSYRCECNEGFEASATGT 2336
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 38/70 (54%), Gaps = 3/70 (4%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
ASC NT GSY C C G++ ++ G C+DV+EC+ G C S C N DG + C C
Sbjct 2627 ASCYNTLGSYKCVCPSGFDFEATAG-GCQDVNECSMGNNPC--SYGCSNTDGGYLCGCPG 2683
Query 87 GFEAADAKTC 96
GF A C
Sbjct 2684 GFYRAGQGHC 2693
Score = 47.8 bits (112), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 36/72 (50%), Gaps = 3/72 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT+GS+ C C GY G C D DEC+ G + C NV G FEC+C
Sbjct 2144 NGLCINTDGSFRCECPFGYN-LDYTGVNCVDTDECSIGNP--CGNGTCSNVPGGFECSCQ 2200
Query 86 AGFEAADAKTCK 97
GFE TC+
Sbjct 2201 EGFEPGPMMTCE 2212
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 35/66 (53%), Gaps = 2/66 (3%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H +C NT GS+ C C+ G+ + + C D+DEC C CVN GSFEC C
Sbjct 1046 HGTCRNTIGSFKCRCDSGFA-LTMEERNCTDIDECTISPDLCG-HGVCVNTPGSFECECF 1103
Query 86 AGFEAA 91
G+E+
Sbjct 1104 DGYESG 1109
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/66 (37%), Positives = 32/66 (48%), Gaps = 1/66 (1%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H C NT GS+ C C GYE C D+DEC C C+N +GS+EC C
Sbjct 1088 HGVCVNTPGSFECECFDGYESGFMMMKNCMDIDECERDPLLCR-GGTCLNTEGSYECDCP 1146
Query 86 AGFEAA 91
G + +
Sbjct 1147 PGHQLS 1152
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 37/67 (55%), Gaps = 4/67 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ +C+N G + C+C G+EP C+D++ECA C + +CVN GS+EC C
Sbjct 2185 NGTCSNVPGGFECSCQEGFEPGPM--MTCEDINECAVNPLLC--AFRCVNTFGSYECMCP 2240
Query 86 AGFEAAD 92
G+ D
Sbjct 2241 TGYVLRD 2247
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/66 (37%), Positives = 33/66 (50%), Gaps = 4/66 (6%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
+C N +GS+ C C PGYE + C D++EC C C N GSF+C C
Sbjct 1990 GTCQNLDGSFRCICPPGYEVQND---KCVDINECNVEPNICQFGT-CKNTPGSFQCICQP 2045
Query 87 GFEAAD 92
GF +D
Sbjct 2046 GFVLSD 2051
Score = 44.7 bits (104), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 40/73 (54%), Gaps = 4/73 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N+ GS+ C CN GY + G +C D+DEC+ C+ C N +GS+ C+C
Sbjct 2422 NGQCINSVGSFRCHCNVGYTNDFT-GTSCIDMDECSQSPKPCNF--LCKNTEGSYLCSCP 2478
Query 86 AGF-EAADAKTCK 97
G+ D KTCK
Sbjct 2479 RGYILQPDGKTCK 2491
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 24/62 (38%), Positives = 35/62 (56%), Gaps = 2/62 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
+C NT GSY C C G+ + ++ C+D+DECA + C +C N GS+ CTC
Sbjct 259 GNCINTVGSYECKCPAGHRQSETN-QKCEDIDECATISGVCD-GGECTNTAGSYVCTCPR 316
Query 87 GF 88
G+
Sbjct 317 GY 318
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 32/70 (45%), Gaps = 5/70 (7%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
L+ C T SY C CN GY C DVDEC + V+ CVN GS+ C C
Sbjct 466 LNGRCIPTPTSYRCECNMGYRQDVRG--ECIDVDECVSNPC---VNGDCVNTQGSYHCKC 520
Query 85 MAGFEAADAK 94
G++ K
Sbjct 521 HEGYQGTPTK 530
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 31/64 (48%), Gaps = 3/64 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N G Y C C G+ P + D AC+D+DEC V C N+ G F C C
Sbjct 1422 NGQCLNAPGGYRCECEMGFTP-TEDSKACQDIDECNFQNI--CVFGSCQNLPGMFRCVCD 1478
Query 86 AGFE 89
G+E
Sbjct 1479 YGYE 1482
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 41/81 (50%), Gaps = 3/81 (3%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E ++ + +A+C N GSY C C+ G++ + S AC D +EC C +C+
Sbjct 1813 ECNSGDNLCQRNANCINIPGSYRCECSAGFKLSPSG--ACIDRNECQEIPNVCS-HGECI 1869
Query 75 NVDGSFECTCMAGFEAADAKT 95
+ GSF C C GF+ +T
Sbjct 1870 DTQGSFRCLCHNGFKTTPDQT 1890
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 26/61 (42%), Positives = 29/61 (47%), Gaps = 2/61 (3%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQ-CVNVDGSFECTCMAG 87
C NT GSY C C GY D C+D DECA G +C C N G+F C C G
Sbjct 2228 CVNTFGSYECMCPTGYV-LRDDNRMCRDQDECAEGLDDCASRGMACKNQIGTFMCICPPG 2286
Query 88 F 88
Sbjct 2287 M 2287
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 35/65 (53%), Gaps = 3/65 (4%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
+ SC N G + C C+ GYE S G+ C D++EC C ++ CVN+ GS+ C C
Sbjct 1462 VFGSCQNLPGMFRCVCDYGYELDRSGGN-CTDINECFD-PVNC-INGVCVNLPGSYLCNC 1518
Query 85 MAGFE 89
FE
Sbjct 1519 PPDFE 1523
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 35/70 (50%), Gaps = 4/70 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT GSY C+C G+ +D C+D++EC + ++ C N+ GSF C C
Sbjct 783 NGMCRNTPGSYTCSCPKGFS-FKADSETCEDINECDSNPC---INGICRNIAGSFNCECS 838
Query 86 AGFEAADAKT 95
G + T
Sbjct 839 HGSKLDSTNT 848
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 29/69 (42%), Gaps = 2/69 (2%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C N G Y C C G+ AS D C DV+EC C + C N GSF C C
Sbjct 1257 GQCTNIPGEYRCLCYDGFM-ASMDMRTCIDVNECDLNPNIC-LHGDCENTKGSFICHCQL 1314
Query 87 GFEAADAKT 95
G+ T
Sbjct 1315 GYFVKKGST 1323
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 35/78 (44%), Gaps = 4/78 (5%)
Query 11 VPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVS 70
+ E S + ASC N+ GS+ C C G+ + G C DVDEC G C
Sbjct 2530 IDNNECSNQPNICGSRASCLNSPGSFNCECQKGFS-LDATGLNCDDVDEC-GGNHRCQHG 2587
Query 71 AQCVNVDGSFECTCMAGF 88
Q N+ G + C C G+
Sbjct 2588 CQ--NMMGGYRCGCPQGY 2603
Score = 37.0 bits (84), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 22/55 (40%), Positives = 28/55 (50%), Gaps = 2/55 (3%)
Query 40 CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADAK 94
C G+ P G AC+DVDEC A C C+N GS+EC C AG ++
Sbjct 230 CRRGFIPNIRTG-ACQDVDECQAVPGLC-AGGNCINTVGSYECKCPAGHRQSETN 282
Score = 35.4 bits (80), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 29/58 (50%), Gaps = 2/58 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
+C NT GS+ C C GY + + C+D+DEC A C C N G++ C C
Sbjct 1624 GNCVNTFGSFQCECPAGYY-LNEETRICEDIDECTAHIGICG-PGTCYNTLGNYTCVC 1679
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 30/61 (49%), Gaps = 3/61 (4%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N G Y C C GY + C D +EC G C SA C N GS++C C +GF
Sbjct 2588 CQNMMGGYRCGCPQGYV-QHYQWNQCVDENECT-GNQVCG-SASCYNTLGSYKCVCPSGF 2644
Query 89 E 89
+
Sbjct 2645 D 2645
Score = 31.6 bits (70), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 24/47 (51%), Gaps = 2/47 (4%)
Query 51 GHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADA-KTC 96
G C+D++EC C + +CVN GSF C C G A +TC
Sbjct 910 GLVCEDINECEVFPGVC-TNGRCVNTQGSFRCECPEGLTLDGAGRTC 955
Score = 31.6 bits (70), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 28/52 (53%), Gaps = 7/52 (13%)
Query 37 VCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
+C PGY ++DG +D++EC C + QC+N GSF C C G+
Sbjct 2396 MCPLGPGY---TTDG---RDINECEVMPNLCK-NGQCINSVGSFRCHCNVGY 2440
Score = 31.2 bits (69), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 17/30 (56%), Positives = 18/30 (60%), Gaps = 1/30 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKD 56
C NT GSYVCTC GY S+DG C D
Sbjct 301 GECTNTAGSYVCTCPRGY-ITSTDGSRCVD 329
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 1/34 (2%)
Query 55 KDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
+D+DEC C CVN GSF+C C AG+
Sbjct 1609 EDIDECQELPGLCQ-GGNCVNTFGSFQCECPAGY 1641
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 18/37 (48%), Gaps = 1/37 (2%)
Query 21 SSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDV 57
S ++ C NTEGS+ C C PG DG C D
Sbjct 625 SGICMNGRCVNTEGSFRCECPPGLA-IDVDGRVCVDT 660
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 16/31 (51%), Positives = 19/31 (61%), Gaps = 1/31 (3%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKD 56
+ C NT GSY C CN G+E AS+ G C D
Sbjct 2311 NGRCVNTVGSYRCECNEGFE-ASATGTECID 2340
Score = 28.9 bits (63), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 18/33 (54%), Gaps = 1/33 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDE 59
+C NT GS+ C C PG+ S +G C D E
Sbjct 2030 GTCKNTPGSFQCICQPGFV-LSDNGRRCFDTRE 2061
> dre:403045 egf; epidermal growth factor; K04357 epidermal growth
factor
Length=1114
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 35/81 (43%), Positives = 46/81 (56%), Gaps = 4/81 (4%)
Query 8 CVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAEC 67
CV V + +A A S S A C NT G Y C C G+ S DGH C D+DEC +C
Sbjct 796 CVDVDECKAGLADCSVS-EAECVNTAGGYFCQCKNGF---SGDGHHCVDIDECRLDLHDC 851
Query 68 HVSAQCVNVDGSFECTCMAGF 88
V+A+C+N G ++C C +GF
Sbjct 852 DVNAECLNAVGEYQCRCRSGF 872
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 31/65 (47%), Positives = 38/65 (58%), Gaps = 4/65 (6%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVS-AQCVNVDGSFECT 83
++A C EG VC C G+ + DG C DVDEC AG A+C VS A+CVN G + C
Sbjct 770 VNAQCFLQEGRAVCQCVRGF---TGDGELCVDVDECKAGLADCSVSEAECVNTAGGYFCQ 826
Query 84 CMAGF 88
C GF
Sbjct 827 CKNGF 831
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 38/70 (54%), Gaps = 4/70 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C++ + +C C G+ S G+ C+DV+EC+ C S C NV GS+ CTC G+
Sbjct 267 CSSHTDTGLCGCKDGFT-LSKHGNICEDVNECSLWNHGC--SLGCENVPGSYFCTCPEGY 323
Query 89 EAA-DAKTCK 97
D KTC+
Sbjct 324 LLLPDLKTCQ 333
> xla:398322 ltbp1, ltgf-1; latent transforming growth factor
beta binding protein 1; K08023 latent transforming growth factor
beta binding protein
Length=1399
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/72 (43%), Positives = 40/72 (55%), Gaps = 1/72 (1%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT GS+ C CN GY+ S+DG C+D+DEC C C N +GS+ECTC
Sbjct 797 NGQCRNTRGSFTCICNQGYK-VSADGEQCEDIDECQEEAEVCLGGGSCHNTEGSYECTCP 855
Query 86 AGFEAADAKTCK 97
GF D C+
Sbjct 856 DGFLLVDGIRCQ 867
Score = 61.6 bits (148), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 35/74 (47%), Positives = 47/74 (63%), Gaps = 4/74 (5%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
L+ C NTEGS+VC+C+ GY+PA S G C D+DEC T C + QC N GSF C C
Sbjct 755 LNGLCQNTEGSFVCSCHEGYQPAPS-GDQCNDIDEC-QNTDLCR-NGQCRNTRGSFTCIC 811
Query 85 MAGFE-AADAKTCK 97
G++ +AD + C+
Sbjct 812 NQGYKVSADGEQCE 825
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 32/74 (43%), Positives = 45/74 (60%), Gaps = 5/74 (6%)
Query 20 HSSTSL--HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVD 77
TSL + C+N EGSY CTC GY+ +SDG C D++EC +GT ++ C N +
Sbjct 707 QEDTSLCANGDCSNLEGSYRCTCQRGYD-VTSDGKECIDINECQSGTL--CLNGLCQNTE 763
Query 78 GSFECTCMAGFEAA 91
GSF C+C G++ A
Sbjct 764 GSFVCSCHEGYQPA 777
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 42/72 (58%), Gaps = 3/72 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NTEGSY C C G+ S++ +C+DVDEC A T +C C N +GS+ C C
Sbjct 881 NGECLNTEGSYHCICEQGFT-TSANERSCEDVDEC-ANTTQCGSQGICENTEGSYRCLCY 938
Query 86 AGFEAA-DAKTC 96
G++ + D + C
Sbjct 939 QGYQDSFDGQGC 950
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 43/72 (59%), Gaps = 4/72 (5%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
SC NTEGSY CTC G+ DG C+D++EC C + +C+N +GS+ C C
Sbjct 841 GSCHNTEGSYECTCPDGF--LLVDGIRCQDINECEQPEL-CTPNGECLNTEGSYHCICEQ 897
Query 87 GF-EAADAKTCK 97
GF +A+ ++C+
Sbjct 898 GFTTSANERSCE 909
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 31/67 (46%), Positives = 36/67 (53%), Gaps = 2/67 (2%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NTEGSY C C GY+ S DG C DV+EC C A+C NV+GSF C C
Sbjct 926 CENTEGSYRCLCYQGYQ-DSFDGQGCVDVNECEMLNGVCG-EARCENVEGSFLCLCADES 983
Query 89 EAADAKT 95
+ D T
Sbjct 984 QEYDPMT 990
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 39/73 (53%), Gaps = 6/73 (8%)
Query 26 HASCANTEGSYVCT-CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
H C N+ GSY C C G+ + C D++EC T+ C + C N++GS+ CTC
Sbjct 674 HDRCVNSVGSYACVPCPDGFNGLNG---QCFDINECQEDTSLC-ANGDCSNLEGSYRCTC 729
Query 85 MAGFEA-ADAKTC 96
G++ +D K C
Sbjct 730 QRGYDVTSDGKEC 742
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 33/73 (45%), Gaps = 3/73 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT-C 84
H C NT+G ++C C+ GY A+ G C DV+EC C N GSF C C
Sbjct 591 HGRCENTKGRFLCICHAGYM-ANEQGTDCVDVNECLR-PHPCEDDGLWFNTIGSFRCEYC 648
Query 85 MAGFEAADAKTCK 97
GF C+
Sbjct 649 EKGFRKNQRGDCE 661
Score = 38.1 bits (87), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 1 LPMVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGH 52
L M CV V + + S +A C NTEGSY C C PGY+P++ +
Sbjct 1333 LDMAKMTCVDVNECNELNSRMSLCKNAKCINTEGSYKCVCLPGYKPSAKPNY 1384
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 35/68 (51%), Gaps = 3/68 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHV--SAQCVNVDGSFECT 83
+ C + Y C C GY + C DV+EC + + +A+C+N +GS++C
Sbjct 1313 NGRCVRVQEGYTCDCFDGYHLDMAK-MTCVDVNECNELNSRMSLCKNAKCINTEGSYKCV 1371
Query 84 CMAGFEAA 91
C+ G++ +
Sbjct 1372 CLPGYKPS 1379
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 31/69 (44%), Gaps = 2/69 (2%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
+C N Y C C GY+ + C D++ECA C +C N G F C C A
Sbjct 550 GNCINLPVGYTCLCFTGYK-LNEQKTKCVDINECAKDPQICS-HGRCENTKGRFLCICHA 607
Query 87 GFEAADAKT 95
G+ A + T
Sbjct 608 GYMANEQGT 616
Score = 35.0 bits (79), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 30/64 (46%), Gaps = 13/64 (20%)
Query 26 HASCANTEGSYVCTCNPG--YEPAS---SDGHACKDVDECAAGTAECHVSAQCVNVDGSF 80
+ C NTE SY C C G Y+P D + C+D C G QC+N +GS+
Sbjct 1124 NGYCLNTESSYECYCKQGTYYDPVKLQCIDNNECEDPSSCIDG--------QCINNEGSY 1175
Query 81 ECTC 84
C C
Sbjct 1176 SCFC 1179
> dre:100332400 novel EGF domain containing protein-like
Length=1932
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 42/72 (58%), Gaps = 8/72 (11%)
Query 22 STSLH-----ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNV 76
+T LH A C NT GSY C+C GY S DG C+D++EC +CH +A C N
Sbjct 17 TTGLHSCHAKAVCTNTLGSYTCSCQNGY---SGDGFQCQDINECQTNNGDCHANALCTNK 73
Query 77 DGSFECTCMAGF 88
DG +C+C +GF
Sbjct 74 DGGRDCSCRSGF 85
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 30/65 (46%), Positives = 40/65 (61%), Gaps = 4/65 (6%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
S +ASC N GSY C+C G++ DG C+DVDEC CHV+A C+N G + C+
Sbjct 308 SPYASCENMPGSYKCSCIAGFK---GDGLVCEDVDECVT-EKRCHVNALCINSPGKYNCS 363
Query 84 CMAGF 88
CM G+
Sbjct 364 CMVGY 368
Score = 58.9 bits (141), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 32/68 (47%), Positives = 39/68 (57%), Gaps = 4/68 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N GS+ CTC GY S +G C D+DEC +G + C AQCVN GS C C++GF
Sbjct 452 CVNNPGSFECTCKEGY---SFNGTKCTDLDECESGVSNCSKFAQCVNTVGSHLCFCLSGF 508
Query 89 EAADAKTC 96
D K C
Sbjct 509 -TGDGKNC 515
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 33/96 (34%), Positives = 50/96 (52%), Gaps = 7/96 (7%)
Query 3 MVFRQ----CVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVD 58
M FR C + + S S LH++C+N GSY C C+ G+ ++ C+D+D
Sbjct 880 MGFRSNGTSCFDIDECSGSQNESICQLHSTCSNIPGSYKCHCDSGFLLNRTE---CQDID 936
Query 59 ECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADAK 94
EC A + C ++ C+N GSF C C +GF+ K
Sbjct 937 ECLADDSPCTANSVCINSVGSFRCLCASGFKEDGLK 972
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 33/88 (37%), Positives = 44/88 (50%), Gaps = 8/88 (9%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E + S+ S A C NT GS++C C G+ + DG C D++EC CH A C
Sbjct 479 ECESGVSNCSKFAQCVNTVGSHLCFCLSGF---TGDGKNCSDINECHFQNGGCHPVASCT 535
Query 75 NVDGSFECTCM-----AGFEAADAKTCK 97
N GSF+CTC +GF+ D C
Sbjct 536 NSPGSFKCTCPLGMTGSGFDCQDVDECN 563
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 35/65 (53%), Gaps = 5/65 (7%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N +G+Y C CN GY+ S+G C D++EC C A C N+ GS+ CTC
Sbjct 152 YKGCKNLQGTYTCLCNSGYQ---SNGQTCVDINECQINF--CSPFADCTNLPGSYRCTCP 206
Query 86 AGFEA 90
GF
Sbjct 207 EGFNG 211
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 29/74 (39%), Positives = 41/74 (55%), Gaps = 2/74 (2%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E S + S ++++C N G Y C+CN G+ S C DVDECA G A+C ++ C
Sbjct 726 ECSVGNFSCPVNSTCYNEVGGYNCSCNNGF--IYSYNSVCLDVDECATGKAQCPNASNCH 783
Query 75 NVDGSFECTCMAGF 88
N GS+ C C G+
Sbjct 784 NTVGSYYCECWDGY 797
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 31/73 (42%), Positives = 39/73 (53%), Gaps = 5/73 (6%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
SL ++C NTEGSY+C C GY +G C D+DEC + C + C N G+F CT
Sbjct 573 SLLSTCHNTEGSYICKCMEGYW---GNGFTCSDLDECFPPSI-CGNNMTCQNFPGTFTCT 628
Query 84 CMAGFEAADAKTC 96
C G D TC
Sbjct 629 CTLGL-VYDLGTC 640
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 47/89 (52%), Gaps = 4/89 (4%)
Query 1 LPMVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDEC 60
L +V+ V + + A ++ +++A C N GS C+C G+ +G CKD+DEC
Sbjct 631 LGLVYDLGTCVTEKDCKNATNACNIYAECKNVHGSNYCSCMKGFH---GNGRDCKDLDEC 687
Query 61 AAGTAECHVSAQCVNVDGSFECTCMAGFE 89
+ C + C N +GSF C C+ G++
Sbjct 688 SQ-IGACPNLSNCFNTEGSFHCDCLQGYQ 715
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 30/71 (42%), Positives = 41/71 (57%), Gaps = 6/71 (8%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
A+C NT GS+ C C GY + C D+DECA G C A C N+ GS++C+C+A
Sbjct 272 AACVNTAGSFFCDCGQGYNFTQ---NQCVDLDECAIGL--CSPYASCENMPGSYKCSCIA 326
Query 87 GFEAADAKTCK 97
GF+ D C+
Sbjct 327 GFK-GDGLVCE 336
Score = 51.2 bits (121), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 28/75 (37%), Positives = 43/75 (57%), Gaps = 6/75 (8%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHV---SAQCVNVDGSFE 81
+A+C N GSYVCTCN GY+ + + C DVDEC+ C C N+ G++
Sbjct 106 WNATCTNNPGSYVCTCNSGYK--GNGNYLCLDVDECSETPGVCSALLGYKGCKNLQGTYT 163
Query 82 CTCMAGFEAADAKTC 96
C C +G++ ++ +TC
Sbjct 164 CLCNSGYQ-SNGQTC 177
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 37/62 (59%), Gaps = 3/62 (4%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
++C NTEGS+ C C GY+ +G C D++EC+ G C V++ C N G + C+C
Sbjct 697 SNCFNTEGSFHCDCLQGYQ---YNGTHCDDINECSVGNFSCPVNSTCYNEVGGYNCSCNN 753
Query 87 GF 88
GF
Sbjct 754 GF 755
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 40/65 (61%), Gaps = 5/65 (7%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAE--CHVSAQCVNVDGSFECT 83
+ +C NT GS+ C C+ G+ S+G +C D+DEC+ E C + + C N+ GS++C
Sbjct 864 NGTCLNTIGSFGCVCDMGFR---SNGTSCFDIDECSGSQNESICQLHSTCSNIPGSYKCH 920
Query 84 CMAGF 88
C +GF
Sbjct 921 CDSGF 925
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/71 (39%), Positives = 40/71 (56%), Gaps = 4/71 (5%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
S A C N GSY CTC G+ + +G AC D++EC + C +A C N+ GS++C+
Sbjct 189 SPFADCTNLPGSYRCTCPEGF---NGNGLACVDINECDRKNS-CDPNALCTNLLGSYKCS 244
Query 84 CMAGFEAADAK 94
C +GF K
Sbjct 245 CRSGFLGIGTK 255
Score = 48.9 bits (115), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 36/67 (53%), Gaps = 3/67 (4%)
Query 25 LHASCANTEGSYVCTCNPGYEPAS---SDGHACKDVDECAAGTAECHVSAQCVNVDGSFE 81
++A C N+ G Y C+C GY + C+D+DEC C + QC N++GS+
Sbjct 349 VNALCINSPGKYNCSCMVGYTGNGFRLVNHTTCQDIDECQLPEKVCGTNEQCTNLEGSYS 408
Query 82 CTCMAGF 88
C C AGF
Sbjct 409 CQCKAGF 415
Score = 48.1 bits (113), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 29/78 (37%), Positives = 34/78 (43%), Gaps = 10/78 (12%)
Query 26 HASCANTEGSYVCTCNPGY---------EPASSDGHACKDVDECAAGTAECHVSAQCVNV 76
+ C N EGSY C C G+ + DG C D+DEC QCVN
Sbjct 397 NEQCTNLEGSYSCQCKAGFSRIIDDFCSDINEWDGLTCSDIDECEENVCP-EKETQCVNN 455
Query 77 DGSFECTCMAGFEAADAK 94
GSFECTC G+ K
Sbjct 456 PGSFECTCKEGYSFNGTK 473
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 38/72 (52%), Gaps = 1/72 (1%)
Query 20 HSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECA-AGTAECHVSAQCVNVDG 78
+S S H+ C NT GS++C C+ G+ + C+D+DEC+ + C+N G
Sbjct 813 NSRCSDHSICVNTLGSFMCLCDDGFTLNGTSDTQCEDIDECSNPDNGSICTNGTCLNTIG 872
Query 79 SFECTCMAGFEA 90
SF C C GF +
Sbjct 873 SFGCVCDMGFRS 884
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 27/65 (41%), Positives = 37/65 (56%), Gaps = 6/65 (9%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTA---ECHVSAQCVNVDGSFECT 83
ASC N+ GS+ CTC G + G C+DVDEC A + C + + C N +GS+ C
Sbjct 532 ASCTNSPGSFKCTCPLG---MTGSGFDCQDVDECNANSTLPHNCSLLSTCHNTEGSYICK 588
Query 84 CMAGF 88
CM G+
Sbjct 589 CMEGY 593
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 32/64 (50%), Gaps = 4/64 (6%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+A C N GSY C+C G+ G C D++ECA C A CVN GSF C C
Sbjct 231 NALCTNLLGSYKCSCRSGFLGI---GTKCTDINECATDNI-CPAVAACVNTAGSFFCDCG 286
Query 86 AGFE 89
G+
Sbjct 287 QGYN 290
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 26/47 (55%), Gaps = 1/47 (2%)
Query 51 GHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADAKTCK 97
G C+D+DEC G CH A C N GS+ C+C G+ + D C+
Sbjct 7 GVQCQDIDECTTGLHSCHAKAVCTNTLGSYTCSCQNGY-SGDGFQCQ 52
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/75 (32%), Positives = 38/75 (50%), Gaps = 4/75 (5%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E A S + ++ C N+ GS+ C C G++ DG C D++EC + C C+
Sbjct 937 ECLADDSPCTANSVCINSVGSFRCLCASGFK---EDGLKCTDINECLS-NGTCRPDQVCI 992
Query 75 NVDGSFECTCMAGFE 89
N GS+ C+C G +
Sbjct 993 NKPGSYLCSCPPGHQ 1007
> dre:100332336 novel EGF domain containing protein-like
Length=1837
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 42/72 (58%), Gaps = 8/72 (11%)
Query 22 STSLH-----ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNV 76
+T LH A C NT GSY C+C GY S DG C+D++EC +CH +A C N
Sbjct 17 TTGLHSCHAKAVCTNTLGSYTCSCQNGY---SGDGFQCQDINECQTNNGDCHANALCTNK 73
Query 77 DGSFECTCMAGF 88
DG +C+C +GF
Sbjct 74 DGGRDCSCRSGF 85
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 30/65 (46%), Positives = 40/65 (61%), Gaps = 4/65 (6%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
S +ASC N GSY C+C G++ DG C+DVDEC CHV+A C+N G + C+
Sbjct 308 SPYASCENMPGSYKCSCIAGFK---GDGLVCEDVDECVT-EKRCHVNALCINSPGKYNCS 363
Query 84 CMAGF 88
CM G+
Sbjct 364 CMVGY 368
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 32/68 (47%), Positives = 39/68 (57%), Gaps = 4/68 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N GS+ CTC GY S +G C D+DEC +G + C AQCVN GS C C++GF
Sbjct 452 CVNNPGSFECTCKEGY---SFNGTKCTDLDECESGVSNCSKFAQCVNTVGSHLCFCLSGF 508
Query 89 EAADAKTC 96
D K C
Sbjct 509 -TGDGKNC 515
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 33/96 (34%), Positives = 50/96 (52%), Gaps = 7/96 (7%)
Query 3 MVFRQ----CVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVD 58
M FR C + + S S LH++C+N GSY C C+ G+ ++ C+D+D
Sbjct 880 MGFRSNGTSCFDIDECSGSQNESICQLHSTCSNIPGSYKCHCDSGFLLNRTE---CQDID 936
Query 59 ECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADAK 94
EC A + C ++ C+N GSF C C +GF+ K
Sbjct 937 ECLADDSPCTANSVCINSVGSFRCLCASGFKEDGLK 972
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 33/88 (37%), Positives = 44/88 (50%), Gaps = 8/88 (9%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E + S+ S A C NT GS++C C G+ + DG C D++EC CH A C
Sbjct 479 ECESGVSNCSKFAQCVNTVGSHLCFCLSGF---TGDGKNCSDINECHFQNGGCHPVASCT 535
Query 75 NVDGSFECTCM-----AGFEAADAKTCK 97
N GSF+CTC +GF+ D C
Sbjct 536 NSPGSFKCTCPLGMTGSGFDCQDVDECN 563
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 35/65 (53%), Gaps = 5/65 (7%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N +G+Y C CN GY+ S+G C D++EC C A C N+ GS+ CTC
Sbjct 152 YKGCKNLQGTYTCLCNSGYQ---SNGQTCVDINECQINF--CSPFADCTNLPGSYRCTCP 206
Query 86 AGFEA 90
GF
Sbjct 207 EGFNG 211
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 29/74 (39%), Positives = 41/74 (55%), Gaps = 2/74 (2%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E S + S ++++C N G Y C+CN G+ S C DVDECA G A+C ++ C
Sbjct 726 ECSVGNFSCPVNSTCYNEVGGYNCSCNNGF--IYSYNSVCLDVDECATGKAQCPNASNCH 783
Query 75 NVDGSFECTCMAGF 88
N GS+ C C G+
Sbjct 784 NTVGSYYCECWDGY 797
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 47/89 (52%), Gaps = 4/89 (4%)
Query 1 LPMVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDEC 60
L +V+ V + + A ++ +++A C N GS C+C G+ +G CKD+DEC
Sbjct 631 LGLVYDLGTCVTEKDCKNATNACNIYAECKNVHGSNYCSCMKGFH---GNGRDCKDLDEC 687
Query 61 AAGTAECHVSAQCVNVDGSFECTCMAGFE 89
+ C + C N +GSF C C+ G++
Sbjct 688 SQ-IGACPNLSNCFNTEGSFHCDCLQGYQ 715
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 31/73 (42%), Positives = 39/73 (53%), Gaps = 5/73 (6%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
SL ++C NTEGSY+C C GY +G C D+DEC + C + C N G+F CT
Sbjct 573 SLLSTCHNTEGSYICKCMEGYW---GNGFTCSDLDECFPPSI-CGNNMTCQNFPGTFTCT 628
Query 84 CMAGFEAADAKTC 96
C G D TC
Sbjct 629 CTLGL-VYDLGTC 640
Score = 51.2 bits (121), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 30/71 (42%), Positives = 41/71 (57%), Gaps = 6/71 (8%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
A+C NT GS+ C C GY + C D+DECA G C A C N+ GS++C+C+A
Sbjct 272 AACVNTAGSFFCDCGQGYNFTQ---NQCVDLDECAIGL--CSPYASCENMPGSYKCSCIA 326
Query 87 GFEAADAKTCK 97
GF+ D C+
Sbjct 327 GFK-GDGLVCE 336
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 28/75 (37%), Positives = 43/75 (57%), Gaps = 6/75 (8%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHV---SAQCVNVDGSFE 81
+A+C N GSYVCTCN GY+ + + C DVDEC+ C C N+ G++
Sbjct 106 WNATCTNNPGSYVCTCNSGYK--GNGNYLCLDVDECSETPGVCSALLGYKGCKNLQGTYT 163
Query 82 CTCMAGFEAADAKTC 96
C C +G++ ++ +TC
Sbjct 164 CLCNSGYQ-SNGQTC 177
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 37/62 (59%), Gaps = 3/62 (4%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
++C NTEGS+ C C GY+ +G C D++EC+ G C V++ C N G + C+C
Sbjct 697 SNCFNTEGSFHCDCLQGYQ---YNGTHCDDINECSVGNFSCPVNSTCYNEVGGYNCSCNN 753
Query 87 GF 88
GF
Sbjct 754 GF 755
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 40/65 (61%), Gaps = 5/65 (7%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAE--CHVSAQCVNVDGSFECT 83
+ +C NT GS+ C C+ G+ S+G +C D+DEC+ E C + + C N+ GS++C
Sbjct 864 NGTCLNTIGSFGCVCDMGFR---SNGTSCFDIDECSGSQNESICQLHSTCSNIPGSYKCH 920
Query 84 CMAGF 88
C +GF
Sbjct 921 CDSGF 925
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 28/71 (39%), Positives = 40/71 (56%), Gaps = 4/71 (5%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
S A C N GSY CTC G+ + +G AC D++EC + C +A C N+ GS++C+
Sbjct 189 SPFADCTNLPGSYRCTCPEGF---NGNGLACVDINECDRKNS-CDPNALCTNLLGSYKCS 244
Query 84 CMAGFEAADAK 94
C +GF K
Sbjct 245 CRSGFLGIGTK 255
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 36/67 (53%), Gaps = 3/67 (4%)
Query 25 LHASCANTEGSYVCTCNPGYEPAS---SDGHACKDVDECAAGTAECHVSAQCVNVDGSFE 81
++A C N+ G Y C+C GY + C+D+DEC C + QC N++GS+
Sbjct 349 VNALCINSPGKYNCSCMVGYTGNGFRLVNHTTCQDIDECQLPEKVCGTNEQCTNLEGSYS 408
Query 82 CTCMAGF 88
C C AGF
Sbjct 409 CQCKAGF 415
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 29/78 (37%), Positives = 34/78 (43%), Gaps = 10/78 (12%)
Query 26 HASCANTEGSYVCTCNPGY---------EPASSDGHACKDVDECAAGTAECHVSAQCVNV 76
+ C N EGSY C C G+ + DG C D+DEC QCVN
Sbjct 397 NEQCTNLEGSYSCQCKAGFSRIIDDFCSDINEWDGLTCSDIDECEENVCP-EKETQCVNN 455
Query 77 DGSFECTCMAGFEAADAK 94
GSFECTC G+ K
Sbjct 456 PGSFECTCKEGYSFNGTK 473
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 38/72 (52%), Gaps = 1/72 (1%)
Query 20 HSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECA-AGTAECHVSAQCVNVDG 78
+S S H+ C NT GS++C C+ G+ + C+D+DEC+ + C+N G
Sbjct 813 NSRCSDHSICVNTLGSFMCLCDDGFTLNGTSDTQCEDIDECSNPDNGSICTNGTCLNTIG 872
Query 79 SFECTCMAGFEA 90
SF C C GF +
Sbjct 873 SFGCVCDMGFRS 884
Score = 47.8 bits (112), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 27/65 (41%), Positives = 37/65 (56%), Gaps = 6/65 (9%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTA---ECHVSAQCVNVDGSFECT 83
ASC N+ GS+ CTC G + G C+DVDEC A + C + + C N +GS+ C
Sbjct 532 ASCTNSPGSFKCTCPLG---MTGSGFDCQDVDECNANSTLPHNCSLLSTCHNTEGSYICK 588
Query 84 CMAGF 88
CM G+
Sbjct 589 CMEGY 593
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 32/64 (50%), Gaps = 4/64 (6%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+A C N GSY C+C G+ G C D++ECA C A CVN GSF C C
Sbjct 231 NALCTNLLGSYKCSCRSGFLGI---GTKCTDINECATDNI-CPAVAACVNTAGSFFCDCG 286
Query 86 AGFE 89
G+
Sbjct 287 QGYN 290
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 26/47 (55%), Gaps = 1/47 (2%)
Query 51 GHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADAKTCK 97
G C+D+DEC G CH A C N GS+ C+C G+ + D C+
Sbjct 7 GVQCQDIDECTTGLHSCHAKAVCTNTLGSYTCSCQNGY-SGDGFQCQ 52
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/75 (32%), Positives = 38/75 (50%), Gaps = 4/75 (5%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E A S + ++ C N+ GS+ C C G++ DG C D++EC + C C+
Sbjct 937 ECLADDSPCTANSVCINSVGSFRCLCASGFK---EDGLKCTDINECLS-NGTCRPDQVCI 992
Query 75 NVDGSFECTCMAGFE 89
N GS+ C+C G +
Sbjct 993 NKPGSYLCSCPPGHQ 1007
> ath:AT1G21240 WAK3; WAK3 (wall associated kinase 3); kinase/
protein serine/threonine kinase
Length=741
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 27/73 (36%), Positives = 39/73 (53%), Gaps = 2/73 (2%)
Query 26 HASCAN--TEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
++SC N T Y+C CN GY+ CKD+DEC + T C C N DG F+C
Sbjct 261 NSSCYNSTTRNGYICKCNEGYDGNPYRSEGCKDIDECISDTHNCSDPKTCRNRDGGFDCK 320
Query 84 CMAGFEAADAKTC 96
C +G++ + +C
Sbjct 321 CPSGYDLNSSMSC 333
> mmu:216616 Efemp1, MGC37612; epidermal growth factor-containing
fibulin-like extracellular matrix protein 1
Length=493
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 31/69 (44%), Positives = 40/69 (57%), Gaps = 3/69 (4%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N GS++C CN GYE SSD C+D+DEC + C QCVN G F C C G+
Sbjct 268 CYNILGSFICQCNQGYE-LSSDRLNCEDIDECRTSSYLCQY--QCVNEPGKFSCMCPQGY 324
Query 89 EAADAKTCK 97
E ++TC+
Sbjct 325 EVVRSRTCQ 333
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 41/69 (59%), Gaps = 4/69 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H C NT GS+ C C+PG++ A+++ + C D++EC A + QC N+ GSF C C
Sbjct 225 HQRCVNTPGSFYCQCSPGFQLAANN-YTCVDINECDASN---QCAQQCYNILGSFICQCN 280
Query 86 AGFEAADAK 94
G+E + +
Sbjct 281 QGYELSSDR 289
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 31/69 (44%), Positives = 37/69 (53%), Gaps = 7/69 (10%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N GS+ C C PGY+ G C D+DEC CH +CVN GSF C C GF
Sbjct 190 CINLRGSFTCQCLPGYQ---KRGEQCVDIDECTV-PPYCH--QRCVNTPGSFYCQCSPGF 243
Query 89 E-AADAKTC 96
+ AA+ TC
Sbjct 244 QLAANNYTC 252
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 22/55 (40%), Positives = 33/55 (60%), Gaps = 2/55 (3%)
Query 35 SYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFE 89
S+ C GYE S+ + C+D+DEC +GT C C+N+ GSF C C+ G++
Sbjct 154 SHRIQCAAGYE--QSEHNVCQDIDECTSGTHNCRTDQVCINLRGSFTCQCLPGYQ 206
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 27/62 (43%), Gaps = 3/62 (4%)
Query 21 SSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSF 80
SS C N G + C C GYE S C+D++EC T EC C N G F
Sbjct 301 SSYLCQYQCVNEPGKFSCMCPQGYEVVRS--RTCQDINECET-TNECREDEMCWNYHGGF 357
Query 81 EC 82
C
Sbjct 358 RC 359
Score = 28.1 bits (61), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 19/43 (44%), Gaps = 1/43 (2%)
Query 40 CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFEC 82
C GYE CKD+DEC C +CVN G + C
Sbjct 29 CTDGYE-WDPIRQQCKDIDECDIVPDACKGGMKCVNHYGGYLC 70
> cel:F56H11.1 hypothetical protein
Length=728
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 32/64 (50%), Positives = 41/64 (64%), Gaps = 3/64 (4%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
L A C NT GS+ C C PG++ A SDG C+DV+EC G A C +CVN+ GS++C C
Sbjct 444 LSAECINTIGSFECKCKPGFQLA-SDGRRCEDVNECTTGIAACE--QKCVNIPGSYQCIC 500
Query 85 MAGF 88
GF
Sbjct 501 DRGF 504
Score = 58.9 bits (141), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 32/71 (45%), Positives = 42/71 (59%), Gaps = 3/71 (4%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAE-CHVSAQCVNVDGSFECTCMAG 87
C N G+Y C C PGYE + C+DVDEC C +SA+C+N GSFEC C G
Sbjct 404 CINLPGTYKCKCGPGYEFNDAK-KRCEDVDECIKFAGHVCDLSAECINTIGSFECKCKPG 462
Query 88 FE-AADAKTCK 97
F+ A+D + C+
Sbjct 463 FQLASDGRRCE 473
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 31/57 (54%), Positives = 36/57 (63%), Gaps = 4/57 (7%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQ-CVNVDGSFEC 82
C NT+GSY+C C PGY+ DG C DVDECA G EC S + CVN GSF+C
Sbjct 532 GGCINTKGSYLCQCPPGYK-IQPDGRTCVDVDECAMG--ECAGSDKVCVNTLGSFKC 585
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 27/72 (37%), Positives = 38/72 (52%), Gaps = 4/72 (5%)
Query 28 SCANTEGSYVCTCNPGYEPASSDGHACKDVDECA--AGTAECHVSAQCVNVDGSFECTCM 85
C N GSY C C+ G+ DG C+D+DEC+ AG+ C+N GS+ C C
Sbjct 488 KCVNIPGSYQCICDRGF-ALGPDGTKCEDIDECSIWAGSGNDLCMGGCINTKGSYLCQCP 546
Query 86 AGFEAA-DAKTC 96
G++ D +TC
Sbjct 547 PGYKIQPDGRTC 558
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 28/71 (39%), Positives = 39/71 (54%), Gaps = 9/71 (12%)
Query 29 CANTEGSYVC-----TCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
C NT GS+ C C GYE + G C+DV+EC G S +C+N+ G+++C
Sbjct 359 CVNTPGSFRCQQKGNLCAHGYEVNGATGF-CEDVNECQQGVCG---SMECINLPGTYKCK 414
Query 84 CMAGFEAADAK 94
C G+E DAK
Sbjct 415 CGPGYEFNDAK 425
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 35/91 (38%), Gaps = 30/91 (32%)
Query 29 CANTEGSYVC---------------------TCNPGYEPASSDGHACKDVDECAAGTAEC 67
C NT+GSY C TC GY P + C D+DEC G C
Sbjct 298 CRNTQGSYRCDAKKCGDGELQNPMTGECTSITCPNGYYPKNG---MCNDIDECVTG-HNC 353
Query 68 HVSAQCVNVDGSFEC-----TCMAGFEAADA 93
+CVN GSF C C G+E A
Sbjct 354 GAGEECVNTPGSFRCQQKGNLCAHGYEVNGA 384
Score = 32.7 bits (73), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 24/45 (53%), Gaps = 1/45 (2%)
Query 38 CTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFEC 82
C+C G++ A DG AC D +EC + C S CVN G + C
Sbjct 178 CSCRSGFDLAP-DGMACVDRNECLTRQSPCTQSEDCVNTIGGYIC 221
Score = 32.0 bits (71), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 23/46 (50%), Gaps = 10/46 (21%)
Query 51 GHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADAKTC 96
GH C DVDEC G+ +C QC N GS+ C DAK C
Sbjct 277 GH-CVDVDECNLGSHDCGPLYQCRNTQGSYRC---------DAKKC 312
> hsa:2015 EMR1, TM7LN3; egf-like module containing, mucin-like,
hormone receptor-like 1; K04591 egf-like module containing,
mucin-like, hormone receptor-like 1
Length=886
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 43/70 (61%), Gaps = 6/70 (8%)
Query 25 LHASCANTEGSYVCTCNPGYEPA------SSDGHACKDVDECAAGTAECHVSAQCVNVDG 78
++++C NT GSY CTC+PG+ P+ + G C+D+DEC + C ++ C N G
Sbjct 231 INSTCTNTPGSYFCTCHPGFAPSNGQLNFTDQGVECRDIDECRQDPSTCGPNSICTNALG 290
Query 79 SFECTCMAGF 88
S+ C C+AGF
Sbjct 291 SYSCGCIAGF 300
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 33/77 (42%), Positives = 46/77 (59%), Gaps = 13/77 (16%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGH--------ACKDVDECAAGTAECHVSAQCVNVD 77
HA+C NT G+Y C CNPG+E SS GH +C+D+DEC T C +++ C N
Sbjct 185 HATCNNTVGNYSCFCNPGFE--SSSGHLSFQGLKASCEDIDEC---TEMCPINSTCTNTP 239
Query 78 GSFECTCMAGFEAADAK 94
GS+ CTC GF ++ +
Sbjct 240 GSYFCTCHPGFAPSNGQ 256
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 39/71 (54%), Gaps = 6/71 (8%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGH------ACKDVDECAAGTAECHVSAQCVNVDGS 79
+A+C NT SY C C G+ ++ H CKD+DEC+ C ++ C N+ G
Sbjct 44 YATCTNTVDSYYCACKQGFLSSNGQNHFKDPGVRCKDIDECSQSPQPCGPNSSCKNLSGR 103
Query 80 FECTCMAGFEA 90
++C+C+ GF +
Sbjct 104 YKCSCLDGFSS 114
Score = 48.1 bits (113), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 28/66 (42%), Positives = 37/66 (56%), Gaps = 4/66 (6%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H+ C N+ GSY C+C G+ S C+DVDECA A C A C N G++ C C
Sbjct 145 HSDCVNSMGSYSCSCQVGF---ISRNSTCEDVDECADPRA-CPEHATCNNTVGNYSCFCN 200
Query 86 AGFEAA 91
GFE++
Sbjct 201 PGFESS 206
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 37/76 (48%), Gaps = 9/76 (11%)
Query 26 HASCANTEGSYVCTCNPGYE--------PASSDGHACKDVDECAAGTAECHVSAQCVNVD 77
++SC N G Y C+C G+ P +C D++EC + C + CVN
Sbjct 94 NSSCKNLSGRYKCSCLDGFSSPTGNDWVPGKPGNFSCTDINECLTSSV-CPEHSDCVNSM 152
Query 78 GSFECTCMAGFEAADA 93
GS+ C+C GF + ++
Sbjct 153 GSYSCSCQVGFISRNS 168
> hsa:8425 LTBP4, FLJ46318, FLJ90018, LTBP-4, LTBP4L, LTBP4S;
latent transforming growth factor beta binding protein 4; K08023
latent transforming growth factor beta binding protein
Length=1624
Score = 61.6 bits (148), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 32/77 (41%), Positives = 39/77 (50%), Gaps = 1/77 (1%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E S+ H C NTEGS+ C+C PGY S C DV+EC G C +C+
Sbjct 837 ECSSGAPPCGPHGHCTNTEGSFRCSCAPGYRAPSGRPGPCADVNECLEGDF-CFPHGECL 895
Query 75 NVDGSFECTCMAGFEAA 91
N DGSF CTC G+
Sbjct 896 NTDGSFACTCAPGYRPG 912
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 32/66 (48%), Positives = 37/66 (56%), Gaps = 3/66 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H C NT+GS+ CTC PGY P G +C DVDEC+ + S C N DGSFEC C
Sbjct 891 HGECLNTDGSFACTCAPGYRPGPR-GASCLDVDECS--EEDLCQSGICTNTDGSFECICP 947
Query 86 AGFEAA 91
G A
Sbjct 948 PGHRAG 953
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 34/71 (47%), Positives = 43/71 (60%), Gaps = 6/71 (8%)
Query 29 CANTEGSYVCT--CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C NT GSY CT C+PGY+P + G C+DVDEC + C A C N+ GSF+C C
Sbjct 1022 CENTPGSYRCTPACDPGYQP--TPGGGCQDVDECR-NRSFCGAHAVCQNLPGSFQCLCDQ 1078
Query 87 GFEAA-DAKTC 96
G+E A D + C
Sbjct 1079 GYEGARDGRHC 1089
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 29/65 (44%), Positives = 37/65 (56%), Gaps = 2/65 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C NT GS++C C GY+ A+ G +C+DVDEC C A C N+ GSF C C A
Sbjct 643 GRCENTPGSFLCVCPAGYQ-AAPHGASCQDVDECTQSPGLCGRGA-CKNLPGSFRCVCPA 700
Query 87 GFEAA 91
GF +
Sbjct 701 GFRGS 705
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 31/70 (44%), Positives = 37/70 (52%), Gaps = 2/70 (2%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
HA C N GS+ C C+ GYE + DG C DV+EC C +A C NV+GSF C C
Sbjct 1062 HAVCQNLPGSFQCLCDQGYE-GARDGRHCVDVNECETLQGVCG-AALCENVEGSFLCVCP 1119
Query 86 AGFEAADAKT 95
E D T
Sbjct 1120 NSPEEFDPMT 1129
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/69 (39%), Positives = 36/69 (52%), Gaps = 2/69 (2%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C NT GS+ C C G+ + G C+DVDECA C +C N +GSF+C C
Sbjct 724 GRCDNTAGSFHCACPAGFR-SRGPGAPCQDVDECARSPPPC-TYGRCENTEGSFQCVCPM 781
Query 87 GFEAADAKT 95
GF+ A +
Sbjct 782 GFQPNTAGS 790
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 27/65 (41%), Positives = 34/65 (52%), Gaps = 2/65 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C N+ GS+ C C PG+ A C DVDEC C + +C N GSF C C A
Sbjct 601 GRCENSPGSFRCVCGPGFR-AGPRAAECLDVDECHRVPPPCDL-GRCENTPGSFLCVCPA 658
Query 87 GFEAA 91
G++AA
Sbjct 659 GYQAA 663
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 34/67 (50%), Gaps = 4/67 (5%)
Query 29 CANTEGSYVC-TCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAG 87
C N+ GS+ C TC G+ C DVDEC++G C C N +GSF C+C G
Sbjct 809 CVNSPGSFQCRTCPSGHHLHRG---RCTDVDECSSGAPPCGPHGHCTNTEGSFRCSCAPG 865
Query 88 FEAADAK 94
+ A +
Sbjct 866 YRAPSGR 872
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 31/71 (43%), Positives = 38/71 (53%), Gaps = 5/71 (7%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECA-AGTAECHVSAQCVNVDGSFECT--CM 85
C NT+GS+ C C PG+ A D +C DVDEC G A C S +C N GS+ C C
Sbjct 935 CTNTDGSFECICPPGHR-AGPDLASCLDVDECRERGPALCG-SQRCENSPGSYRCVRDCD 992
Query 86 AGFEAADAKTC 96
G+ A TC
Sbjct 993 PGYHAGPEGTC 1003
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/66 (42%), Positives = 33/66 (50%), Gaps = 7/66 (10%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACK-DVDECAAGTAECHVSAQCVNVDGSFECTC 84
+C N GS+ C C PA G AC+ DVDECA C +C N GSF C C
Sbjct 684 RGACKNLPGSFRCVC-----PAGFRGSACEEDVDECAQEPPPCG-PGRCDNTAGSFHCAC 737
Query 85 MAGFEA 90
AGF +
Sbjct 738 PAGFRS 743
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 28/65 (43%), Gaps = 2/65 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C + Y C C+ G+ S G C DVDEC C +C N GSF C C
Sbjct 559 GRCISRPSGYTCACDSGFR-LSPQGTRCIDVDECRRVPPPC-APGRCENSPGSFRCVCGP 616
Query 87 GFEAA 91
GF A
Sbjct 617 GFRAG 621
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 37/73 (50%), Gaps = 6/73 (8%)
Query 29 CANTEGSYVCT--CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT--C 84
C N+ GSY C C+PGY A +G C DVDEC E + +C N GS+ CT C
Sbjct 978 CENSPGSYRCVRDCDPGYH-AGPEG-TCDDVDECQEYGPEICGAQRCENTPGSYRCTPAC 1035
Query 85 MAGFEAADAKTCK 97
G++ C+
Sbjct 1036 DPGYQPTPGGGCQ 1048
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 31/65 (47%), Gaps = 3/65 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDEC--AAGTAECHVSAQCVNVDGSFECT 83
+ C + C C GY AC D++EC A + V+A+C+N DGSF C
Sbjct 1545 NGRCVRVPEGFTCRCFDGYR-LDMTRMACVDINECDEAEAASPLCVNARCLNTDGSFRCI 1603
Query 84 CMAGF 88
C GF
Sbjct 1604 CRPGF 1608
Score = 35.8 bits (81), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 1 LPMVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGH 52
L M CV + + + + A S ++A C NT+GS+ C C PG+ P H
Sbjct 1565 LDMTRMACVDINECDEAEAASPLCVNARCLNTDGSFRCICRPGFAPTHQPHH 1616
Score = 34.3 bits (77), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 23/56 (41%), Positives = 27/56 (48%), Gaps = 2/56 (3%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
C NT Y C C+ GY + C D DECA C +CVN GS+ CTC
Sbjct 1270 CVNTAPGYSCYCSNGYY-YHTQRLECIDNDECADEEPACE-GGRCVNTVGSYHCTC 1323
Score = 31.6 bits (70), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 20/36 (55%), Gaps = 2/36 (5%)
Query 53 ACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
+C+DVDECA G H +C N G + C C GF
Sbjct 354 SCEDVDECATGGRCQH--GECANTRGGYTCVCPDGF 387
> cel:C50H2.3 mec-9; MEChanosensory abnormality family member
(mec-9)
Length=838
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 33/83 (39%), Positives = 46/83 (55%), Gaps = 7/83 (8%)
Query 6 RQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTA 65
+ C +V G +S S+HA+C N VC C GYE DG C DV+EC
Sbjct 236 KHCTYVGLGRSSIDCKDCSMHATCMNG----VCQCKEGYE---GDGFNCTDVNECLRRPE 288
Query 66 ECHVSAQCVNVDGSFECTCMAGF 88
C+ +A+C+N +GSF CTC+ G+
Sbjct 289 MCNKNAECINREGSFICTCLEGY 311
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 36/67 (53%), Gaps = 3/67 (4%)
Query 30 ANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFE 89
+N++ +Y C C GYE C D+DEC A C +A CVN GS++C CMA +
Sbjct 175 SNSKFNYTCECRSGYEKNQYG--ECIDIDECRGYKAVCDRNAWCVNEIGSYKCECMASYR 232
Query 90 AADAKTC 96
D K C
Sbjct 233 -GDGKHC 238
> hsa:2202 EFEMP1, DHRD, DRAD, FBLN3, FBNL, FLJ35535, MGC111353,
MLVT, MTLV, S1-5; EGF containing fibulin-like extracellular
matrix protein 1
Length=493
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 30/69 (43%), Positives = 40/69 (57%), Gaps = 3/69 (4%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N GS++C CN GYE SSD C+D+DEC + C QCVN G F C C G+
Sbjct 268 CYNILGSFICQCNQGYE-LSSDRLNCEDIDECRTSSYLCQY--QCVNEPGKFSCMCPQGY 324
Query 89 EAADAKTCK 97
+ ++TC+
Sbjct 325 QVVRSRTCQ 333
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 41/69 (59%), Gaps = 4/69 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H C NT GS+ C C+PG++ A+++ + C D++EC A + QC N+ GSF C C
Sbjct 225 HQRCVNTPGSFYCQCSPGFQLAANN-YTCVDINECDASN---QCAQQCYNILGSFICQCN 280
Query 86 AGFEAADAK 94
G+E + +
Sbjct 281 QGYELSSDR 289
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 31/69 (44%), Positives = 37/69 (53%), Gaps = 7/69 (10%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N GS+ C C PGY+ G C D+DEC CH +CVN GSF C C GF
Sbjct 190 CINLRGSFACQCPPGYQ---KRGEQCVDIDECTI-PPYCH--QRCVNTPGSFYCQCSPGF 243
Query 89 E-AADAKTC 96
+ AA+ TC
Sbjct 244 QLAANNYTC 252
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 23/55 (41%), Positives = 32/55 (58%), Gaps = 2/55 (3%)
Query 35 SYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFE 89
S+ C GYE S+ + C+D+DEC AGT C C+N+ GSF C C G++
Sbjct 154 SHRIQCAAGYE--QSEHNVCQDIDECTAGTHNCRADQVCINLRGSFACQCPPGYQ 206
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 27/62 (43%), Gaps = 3/62 (4%)
Query 21 SSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSF 80
SS C N G + C C GY+ S C+D++EC T EC C N G F
Sbjct 301 SSYLCQYQCVNEPGKFSCMCPQGYQVVRS--RTCQDINECET-TNECREDEMCWNYHGGF 357
Query 81 EC 82
C
Sbjct 358 RC 359
Score = 27.7 bits (60), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 19/43 (44%), Gaps = 1/43 (2%)
Query 40 CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFEC 82
C GYE CKD+DEC C +CVN G + C
Sbjct 29 CTDGYE-WDPVRQQCKDIDECDIVPDACKGGMKCVNHYGGYLC 70
> cel:ZK783.1 fbn-1; FiBrilliN homolog family member (fbn-1)
Length=2585
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 31/73 (42%), Positives = 43/73 (58%), Gaps = 4/73 (5%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
SL+A+C N G++ C+C GY DG C D++EC CH A+C N++GSF+C
Sbjct 1596 SLNANCVNMNGTFSCSCKQGYR---GDGFMCTDINECDE-RHPCHPHAECTNLEGSFKCE 1651
Query 84 CMAGFEAADAKTC 96
C +GFE K C
Sbjct 1652 CHSGFEGDGIKKC 1664
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/70 (50%), Positives = 39/70 (55%), Gaps = 4/70 (5%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
A C NTEGSY C C GYE +G C D+DEC G A C A C+N GS C CMA
Sbjct 1734 AVCVNTEGSYRCECAEGYE---GEGGVCTDIDECDRGMAGCDSMAMCINRMGSCGCKCMA 1790
Query 87 GFEAADAKTC 96
G+ D TC
Sbjct 1791 GY-TGDGATC 1799
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 29/65 (44%), Positives = 40/65 (61%), Gaps = 4/65 (6%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAG-TAECHVSAQCVNVDGSFECT 83
+ A C NT GSY C C PGYE +GH C D+DEC+ T+ C ++C+N+ G++ C
Sbjct 132 MMAQCQNTLGSYECRCLPGYE---GNGHECTDIDECSDKLTSRCPEHSKCINLPGTYYCN 188
Query 84 CMAGF 88
C GF
Sbjct 189 CTQGF 193
Score = 55.8 bits (133), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 33/73 (45%), Positives = 39/73 (53%), Gaps = 3/73 (4%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
L A C G VC C PG+E A +C DVDECA G CH SA+C N G + C C
Sbjct 1422 LDALCERRTG--VCRCEPGFEGAPPK-KSCVDVDECATGDHNCHESARCQNYVGGYACFC 1478
Query 85 MAGFEAADAKTCK 97
GF AD +C+
Sbjct 1479 PTGFRKADDGSCQ 1491
Score = 50.8 bits (120), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 35/74 (47%), Gaps = 5/74 (6%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHA---CKDVDECAAGTAECHVSAQCVNVDGSFEC 82
H+ C N G+Y C C G+ P + G C D++EC G C C N GSF+C
Sbjct 175 HSKCINLPGTYYCNCTQGFTPKGNQGSGLDKCADINECETGAHNCDADEICENSIGSFKC 234
Query 83 T--CMAGFEAADAK 94
C G+E D K
Sbjct 235 VNKCSPGYELIDGK 248
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 45/78 (57%), Gaps = 5/78 (6%)
Query 15 EASAAH-SSTSLHASCANTEGSYVCT-CNPGYEPASSDGHACKDVDECAAGTAECHVSAQ 72
++S +H S +++ +GS C C GY+ + G C+D++EC A A C ++A
Sbjct 1544 QSSKSHCSESNMSCEVDTVDGSVECKECMGGYKKS---GKVCEDINECVAEKAPCSLNAN 1600
Query 73 CVNVDGSFECTCMAGFEA 90
CVN++G+F C+C G+
Sbjct 1601 CVNMNGTFSCSCKQGYRG 1618
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 25/63 (39%), Positives = 34/63 (53%), Gaps = 3/63 (4%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAEC-HVSAQCVNVDGSFECTCM 85
A C N G Y C C G+ A D +C+D+DEC + C +A+CVN G++ C C
Sbjct 1465 ARCQNYVGGYACFCPTGFRKA--DDGSCQDIDECTEHNSTCCGANAKCVNKPGTYSCECE 1522
Query 86 AGF 88
GF
Sbjct 1523 NGF 1525
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 33/67 (49%), Gaps = 2/67 (2%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECH-VSAQCVNVDGSFECT 83
L N S VC C PG+ + ++C D+DEC C SA CVN +GS+ C
Sbjct 1688 LSVRIYNGSLSSVCECEPGFR-FEKESNSCVDIDECEESRNNCDPASAVCVNTEGSYRCE 1746
Query 84 CMAGFEA 90
C G+E
Sbjct 1747 CAEGYEG 1753
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 34/70 (48%), Gaps = 4/70 (5%)
Query 26 HASCANTE-GSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
HA C + S+ C+C G+ DG C DVDEC + +C N GSF C C
Sbjct 1870 HAECIDIHPDSHFCSCPDGF---IGDGMICDDVDECNNAGMCDDENTKCENTIGSFNCVC 1926
Query 85 MAGFEAADAK 94
+ GF+ D K
Sbjct 1927 LEGFKKVDEK 1936
Score = 37.4 bits (85), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 25/46 (54%), Gaps = 3/46 (6%)
Query 37 VCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFEC 82
+C C GY + DG C D++EC A C +C+N+DG + C
Sbjct 326 ICDCKTGY---TGDGITCHDINECDAKDTPCSDGGRCLNLDGGYVC 368
Score = 36.2 bits (82), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 28/53 (52%), Gaps = 5/53 (9%)
Query 38 CTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEA 90
C C+ G+ G AC+ D+ CH+ AQC N GS+EC C+ G+E
Sbjct 106 CYCDSGFS-----GSACELQDKNECLEHPCHMMAQCQNTLGSYECRCLPGYEG 153
Score = 35.4 bits (80), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 24/55 (43%), Positives = 32/55 (58%), Gaps = 6/55 (10%)
Query 29 CANTEGSYVCT--CNPGYEPASSDGHACKDVDECAAGTA-ECHVSAQCVNVDGSF 80
C N+ GS+ C C+PGYE DG C+DV+EC + +C V A CVN G +
Sbjct 225 CENSIGSFKCVNKCSPGYELI--DGK-CEDVNECGSEKLHKCDVRADCVNTIGGY 276
Score = 34.7 bits (78), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 28/61 (45%), Gaps = 3/61 (4%)
Query 38 CTCNPGYEPAS--SDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADAKT 95
C C G+ S S C D+DECA + +C A C N GS CTC G D T
Sbjct 2291 CICKSGFRRNSTLSGSETCADIDECAEKSHKCDRVATCRNTFGSHVCTCPDGH-VGDGIT 2349
Query 96 C 96
C
Sbjct 2350 C 2350
Score = 32.0 bits (71), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 43/89 (48%), Gaps = 15/89 (16%)
Query 20 HSST--SLHASCANTEGSYVCTCNPGYEPASSDGHAC-----KDVDECAAGTAEC---HV 69
H+ST +A C N G+Y C C G+ DG+ C K D + + C ++
Sbjct 1499 HNSTCCGANAKCVNKPGTYSCECENGF---LGDGYQCVPTTKKPCDSTQSSKSHCSESNM 1555
Query 70 SAQCVNVDGSFECT-CMAGFEAADAKTCK 97
S + VDGS EC CM G++ + K C+
Sbjct 1556 SCEVDTVDGSVECKECMGGYKKS-GKVCE 1583
> mmu:13733 Emr1, DD7A5-7, EGF-TM7, F4/80, Gpf480, Ly71, TM7LN3;
EGF-like module containing, mucin-like, hormone receptor-like
sequence 1; K04591 egf-like module containing, mucin-like,
hormone receptor-like 1
Length=931
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 31/73 (42%), Positives = 45/73 (61%), Gaps = 10/73 (13%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGH--------ACKDVDECAAGTAECHVSAQCVNV 76
L++SC NT GSY CTC+PG+ ASS+G C+D+DEC +C +++ C NV
Sbjct 282 LNSSCTNTIGSYFCTCHPGF--ASSNGQLNFKDLEVTCEDIDECTQDPLQCGLNSVCTNV 339
Query 77 DGSFECTCMAGFE 89
GS+ C C+ F+
Sbjct 340 PGSYICGCLPDFQ 352
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 33/71 (46%), Positives = 42/71 (59%), Gaps = 10/71 (14%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDG--------HACKDVDECAAGTAECHVSAQCVNVD 77
HA+C NT GSY CTCN G E SS G +C+DVDEC+ + C + C+N
Sbjct 186 HATCHNTLGSYYCTCNSGLE--SSGGGPMFQGLDESCEDVDECSRNSTLCGPTFICINTL 243
Query 78 GSFECTCMAGF 88
GS+ C+C AGF
Sbjct 244 GSYSCSCPAGF 254
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 39/66 (59%), Gaps = 4/66 (6%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+++C+N+ GSY CTC PG+ +G C+D DEC C A C N GS+ CTC
Sbjct 146 YSNCSNSVGSYSCTCQPGF---VLNGSICEDEDECVTRDV-CPEHATCHNTLGSYYCTCN 201
Query 86 AGFEAA 91
+G E++
Sbjct 202 SGLESS 207
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 24/75 (32%), Positives = 41/75 (54%), Gaps = 6/75 (8%)
Query 26 HASCANTEGSYVCTCNPGYEPASSD------GHACKDVDECAAGTAECHVSAQCVNVDGS 79
+A+C +T SY CTC G+ ++ G C+DV+EC + C ++ C N+ G
Sbjct 45 YATCTDTTDSYYCTCKRGFLSSNGQTNFQGPGVECQDVNECLQSDSPCGPNSVCTNILGR 104
Query 80 FECTCMAGFEAADAK 94
+C+C+ GF ++ K
Sbjct 105 AKCSCLRGFSSSTGK 119
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 40/73 (54%), Gaps = 11/73 (15%)
Query 29 CANTEGSYVCTCNPGYEPAS-------SDGHACKDVDECAAGTAECHVSAQCVNVDGSFE 81
C NT GSY C+C G+ + +DG+ C D+DEC C +++ C N GS+
Sbjct 239 CINTLGSYSCSCPAGFSLPTFQILGHPADGN-CTDIDEC---DDTCPLNSSCTNTIGSYF 294
Query 82 CTCMAGFEAADAK 94
CTC GF +++ +
Sbjct 295 CTCHPGFASSNGQ 307
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 31/71 (43%), Gaps = 9/71 (12%)
Query 26 HASCANTEGSYVCTCNPGYEPA--------SSDGHACKDVDECAAGTAECHVSAQCVNVD 77
++ C N G C+C G+ + S D C DVDEC C + C N
Sbjct 95 NSVCTNILGRAKCSCLRGFSSSTGKDWILGSLDNFLCADVDECLT-IGICPKYSNCSNSV 153
Query 78 GSFECTCMAGF 88
GS+ CTC GF
Sbjct 154 GSYSCTCQPGF 164
Score = 28.1 bits (61), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 24/53 (45%), Gaps = 1/53 (1%)
Query 43 GYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADAKT 95
G + G V+EC T C A C + S+ CTC GF +++ +T
Sbjct 19 GMTTLPTLGQTLGGVNECQ-DTTTCPAYATCTDTTDSYYCTCKRGFLSSNGQT 70
> dre:436730 egfl6, sb:cb3, wu:fc66b04, zgc:92328; EGF-like-domain,
multiple 6
Length=508
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 28/68 (41%), Positives = 40/68 (58%), Gaps = 1/68 (1%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDG-HACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
+ C NT GSY C C PGY+ +G + C DV+EC + T +C A+C+N GS++C
Sbjct 153 FNRQCTNTFGSYYCKCQPGYDLKYINGKYDCVDVNECTSNTHKCSHHAECINTLGSYKCK 212
Query 84 CMAGFEAA 91
C GF +
Sbjct 213 CKQGFRGS 220
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/84 (32%), Positives = 43/84 (51%), Gaps = 4/84 (4%)
Query 9 VFVPQGEASAAHSSTSLH--ASCANTEGSYVCTC-NPGYEPASSDGHACKDVDECAAGTA 65
+ +P G + + + + H C + C C +PG + SDG C+D+DECA G
Sbjct 91 MLMPDGSCANSRTCSLAHCQYGCEEVQSEVRCLCPSPGLQ-LGSDGKTCEDIDECATGKN 149
Query 66 ECHVSAQCVNVDGSFECTCMAGFE 89
+C + QC N GS+ C C G++
Sbjct 150 QCPFNRQCTNTFGSYYCKCQPGYD 173
Score = 31.2 bits (69), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 35/78 (44%), Gaps = 17/78 (21%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHAC-KDVDECAAGTAECHVSAQCVNVDGSFECTC 84
H C G C C PGY G C +D++EC C +C+N GS+ C C
Sbjct 37 HGECV---GPNKCKCFPGYT-----GKTCSQDLNECGLKPRPCE--HRCMNTFGSYMCYC 86
Query 85 MAGFEA------ADAKTC 96
+ G+ A+++TC
Sbjct 87 LNGYMLMPDGSCANSRTC 104
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 15/27 (55%), Gaps = 0/27 (0%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSD 50
S HA C NT GSY C C G+ + D
Sbjct 197 SHHAECINTLGSYKCKCKQGFRGSGFD 223
> hsa:255743 NPNT, EGFL6L, POEM; nephronectin; K06824 nephronectin
Length=565
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 34/92 (36%), Positives = 50/92 (54%), Gaps = 4/92 (4%)
Query 6 RQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDG-HACKDVDECAAGT 64
R CV V E + +S C NT GSY+C C+ G++ G + C D+DEC+ G
Sbjct 165 RTCVDV--DECATGRASCPRFRQCVNTFGSYICKCHKGFDLMYIGGKYQCHDIDECSLGQ 222
Query 65 AECHVSAQCVNVDGSFECTCMAGFEAADAKTC 96
+C A+C N+ GS++C C G++ D TC
Sbjct 223 YQCSSFARCYNIRGSYKCKCKEGYQ-GDGLTC 253
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 34/84 (40%), Positives = 43/84 (51%), Gaps = 4/84 (4%)
Query 9 VFVPQGEASAAH--SSTSLHASCANTEGSYVCTC-NPGYEPASSDGHACKDVDECAAGTA 65
+ +P G S+A S + C +G C C +PG + A DG C DVDECA G A
Sbjct 120 MLMPDGSCSSALTCSMANCQYGCDVVKGQIRCQCPSPGLQLAP-DGRTCVDVDECATGRA 178
Query 66 ECHVSAQCVNVDGSFECTCMAGFE 89
C QCVN GS+ C C GF+
Sbjct 179 SCPRFRQCVNTFGSYICKCHKGFD 202
Score = 33.5 bits (75), Expect = 0.18, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 8/64 (12%)
Query 34 GSYVCTCNPGYEPASSDGHAC-KDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAAD 92
G C C+PGY G C +D++EC C +C+N GS++C C+ G+
Sbjct 71 GPNKCKCHPGYA-----GKTCNQDLNECGLKPRPC--KHRCMNTYGSYKCYCLNGYMLMP 123
Query 93 AKTC 96
+C
Sbjct 124 DGSC 127
> hsa:83872 HMCN1, ARMD1, FBLN6, FIBL-6, FIBL6; hemicentin 1
Length=5635
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 30/73 (41%), Positives = 40/73 (54%), Gaps = 4/73 (5%)
Query 28 SCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT--CM 85
SC N G+Y C+C G A+ DG C+D+DECA G CH C N GS+ C C
Sbjct 5120 SCHNAMGTYYCSCPKGLTIAA-DGRTCQDIDECALGRHTCHAGQDCDNTIGSYRCVVRCG 5178
Query 86 AGF-EAADAKTCK 97
+GF +D +C+
Sbjct 5179 SGFRRTSDGLSCQ 5191
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 23/54 (42%), Positives = 32/54 (59%), Gaps = 1/54 (1%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFEC 82
C NT GSY C C PGY+ + +G C+D+DEC C + C N+ GS++C
Sbjct 5446 CKNTFGSYQCICPPGYQ-LTHNGKTCQDIDECLEQNVHCGPNRMCFNMRGSYQC 5498
Score = 51.6 bits (122), Expect = 6e-07, Method: Composition-based stats.
Identities = 28/69 (40%), Positives = 34/69 (49%), Gaps = 4/69 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT GSY C C GY + G C D++EC C + QC N GSF+C C G
Sbjct 5289 CENTRGSYRCVCPRGYR-SQGVGRPCMDINECEQVPKPC--AHQCSNTPGSFKCICPPGQ 5345
Query 89 E-AADAKTC 96
D K+C
Sbjct 5346 HLLGDGKSC 5354
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 26/63 (41%), Positives = 37/63 (58%), Gaps = 6/63 (9%)
Query 29 CANTEGSYVCT--CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C NT GSY C C G+ +SDG +C+D++EC ++ CH +C N GSF C C
Sbjct 5164 CDNTIGSYRCVVRCGSGFR-RTSDGLSCQDINECQE-SSPCH--QRCFNAIGSFHCGCEP 5219
Query 87 GFE 89
G++
Sbjct 5220 GYQ 5222
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 27/79 (34%), Positives = 34/79 (43%), Gaps = 7/79 (8%)
Query 20 HSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGS 79
S+ H C N GS+ C C PGY+ G C DV+EC C C N G
Sbjct 5197 QESSPCHQRCFNAIGSFHCGCEPGYQ---LKGRKCMDVNECRQNV--CRPDQHCKNTRGG 5251
Query 80 FECT--CMAGFEAADAKTC 96
++C C G A+ TC
Sbjct 5252 YKCIDLCPNGMTKAENGTC 5270
Score = 48.1 bits (113), Expect = 7e-06, Method: Composition-based stats.
Identities = 24/64 (37%), Positives = 31/64 (48%), Gaps = 4/64 (6%)
Query 29 CANTEGSYVCT--CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C NT G Y C C G A + C D+DEC GT +C + C N GS+ C C
Sbjct 5245 CKNTRGGYKCIDLCPNGMTKAENG--TCIDIDECKDGTHQCRYNQICENTRGSYRCVCPR 5302
Query 87 GFEA 90
G+ +
Sbjct 5303 GYRS 5306
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 28/74 (37%), Positives = 36/74 (48%), Gaps = 7/74 (9%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
+HAS + + S C P S G C D DECAAG H C N G++ C+C
Sbjct 5079 IHASISKGDRSNQC---PSGFTLDSVGPFCADEDECAAGNPCSH---SCHNAMGTYYCSC 5132
Query 85 MAGFE-AADAKTCK 97
G AAD +TC+
Sbjct 5133 PKGLTIAADGRTCQ 5146
Score = 37.0 bits (84), Expect = 0.016, Method: Composition-based stats.
Identities = 31/94 (32%), Positives = 45/94 (47%), Gaps = 11/94 (11%)
Query 5 FRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGT 64
+RQ + + +S TSL + T + TC G E AS D C D+DEC
Sbjct 5388 YRQYSHLYSSYSEYRNSRTSL----SRTRRTIRKTCPEGSE-ASHD--TCVDIDECENTD 5440
Query 65 AECHVSAQCVNVDGSFECTCMAGFEAA-DAKTCK 97
A H +C N GS++C C G++ + KTC+
Sbjct 5441 ACQH---ECKNTFGSYQCICPPGYQLTHNGKTCQ 5471
Score = 28.5 bits (62), Expect = 6.0, Method: Composition-based stats.
Identities = 12/30 (40%), Positives = 18/30 (60%), Gaps = 1/30 (3%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVD 58
C+NT GS+ C C PG + DG +C ++
Sbjct 5330 CSNTPGSFKCICPPG-QHLLGDGKSCAGLE 5358
> xla:380046 egfl6, MGC53438; EGF-like-domain, multiple 6
Length=544
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 30/77 (38%), Positives = 44/77 (57%), Gaps = 1/77 (1%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDG-HACKDVDECAAGTAECHVSAQC 73
E + +S ++ C NT GSY C C GYE +G + C D++EC T +C ++A C
Sbjct 183 ECAVGKASCPINRRCVNTFGSYYCKCQIGYELKYVNGRYDCIDINECLLNTHKCSINADC 242
Query 74 VNVDGSFECTCMAGFEA 90
+N GSF+C C GF+
Sbjct 243 LNTQGSFKCRCKHGFKG 259
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/83 (33%), Positives = 40/83 (48%), Gaps = 2/83 (2%)
Query 9 VFVPQGEASAAHSST--SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAE 66
+ +P G S + + + C +G C C G DG C D+DECA G A
Sbjct 131 MLMPDGSCSNSRTCAMANCQYGCEQVKGDIRCLCPSGGLQLGPDGRTCIDIDECAVGKAS 190
Query 67 CHVSAQCVNVDGSFECTCMAGFE 89
C ++ +CVN GS+ C C G+E
Sbjct 191 CPINRRCVNTFGSYYCKCQIGYE 213
Score = 32.0 bits (71), Expect = 0.45, Method: Composition-based stats.
Identities = 21/84 (25%), Positives = 36/84 (42%), Gaps = 9/84 (10%)
Query 13 QGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQ 72
+G+ A H C G C C PG+ + + +D++EC C +
Sbjct 64 KGQCEAVCEQGCKHGECV---GPNKCKCFPGFTGKNCN----QDLNECGLKPRPCE--HR 114
Query 73 CVNVDGSFECTCMAGFEAADAKTC 96
C+N GS++C C+ G+ +C
Sbjct 115 CMNTHGSYKCYCLNGYMLMPDGSC 138
> dre:100333499 latent transforming growth factor beta binding
protein 2-like; K08023 latent transforming growth factor beta
binding protein
Length=1089
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 45/73 (61%), Gaps = 3/73 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT+GSY C C PGY A + G C+D++EC G C +CVN DGS++C C
Sbjct 978 QSVCVNTDGSYRCDCAPGYRAAGA-GRQCRDINECLEGDF-CFPRGECVNTDGSYKCVCS 1035
Query 86 AGFEA-ADAKTCK 97
G+++ A+ +C+
Sbjct 1036 QGYKSVANGTSCQ 1048
Score = 48.1 bits (113), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 28/66 (42%), Positives = 36/66 (54%), Gaps = 5/66 (7%)
Query 29 CANTEGSYVC-TCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAG 87
C N+ GS+ C +C PG++ DG C+DVDEC+ C + QC N GSF C C G
Sbjct 833 CVNSVGSFKCVSCKPGFQLL--DGQ-CQDVDECSRTPRRC-TNGQCKNTPGSFRCVCDVG 888
Query 88 FEAADA 93
F D
Sbjct 889 FHLQDG 894
Score = 47.8 bits (112), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 34/64 (53%), Gaps = 2/64 (3%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT GS+ C C GY+ +S C D+DEC C ++ +C N GSF C C G+
Sbjct 537 CVNTPGSFRCVCPAGYQ-TNSQQTLCVDLDECRQVPNPC-INGRCENTLGSFRCVCRTGY 594
Query 89 EAAD 92
+ D
Sbjct 595 KLQD 598
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/71 (39%), Positives = 41/71 (57%), Gaps = 6/71 (8%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFEC-T 83
++ C NT GSY C+C PG+ + + CKDV+EC +C +CVN GSF+C +
Sbjct 790 VNGRCENTMGSYSCSCRPGFR---LEENICKDVNEC-ENELQCP-GMECVNSVGSFKCVS 844
Query 84 CMAGFEAADAK 94
C GF+ D +
Sbjct 845 CKPGFQLLDGQ 855
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 39/66 (59%), Gaps = 3/66 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFEC--T 83
C NT+GSY C C+ GY+ + ++G +C+DV+EC T S +C N GSF C +
Sbjct 1020 RGECVNTDGSYKCVCSQGYK-SVANGTSCQDVNECQDSTGLICGSQRCENTIGSFHCVAS 1078
Query 84 CMAGFE 89
C G++
Sbjct 1079 CEPGYQ 1084
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 24/62 (38%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Query 34 GSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADA 93
G + C CN G+ GH C+D +EC C V +CVN GSF C C AG++
Sbjct 500 GRHTCVCNTGFILNPQQGH-CQDKNECVLTPRPCSV-GECVNTPGSFRCVCPAGYQTNSQ 557
Query 94 KT 95
+T
Sbjct 558 QT 559
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 37/65 (56%), Gaps = 5/65 (7%)
Query 29 CANTEGSYVC-TCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAG 87
C N++GSY C +C PG+ + AC D+DEC T + V+ +C N GS+ C+C G
Sbjct 753 CLNSQGSYRCVSCRPGFGLRNG---ACSDIDECRQ-TPDLCVNGRCENTMGSYSCSCRPG 808
Query 88 FEAAD 92
F +
Sbjct 809 FRLEE 813
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 37/71 (52%), Gaps = 6/71 (8%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFEC-T 83
++ C NT GS+ C C GY+ ++C D+DEC C +CVN GS+ C +
Sbjct 575 INGRCENTLGSFRCVCRTGYK---LQDNSCIDIDEC-VDPLRCP-GKECVNSQGSYRCVS 629
Query 84 CMAGFEAADAK 94
C GF+ + +
Sbjct 630 CRPGFDLLNGQ 640
Score = 34.7 bits (78), Expect = 0.071, Method: Compositional matrix adjust.
Identities = 22/57 (38%), Positives = 31/57 (54%), Gaps = 5/57 (8%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFEC 82
+ C NT GS+ C C+ G+ DG C D+DEC +C QC+N GS++C
Sbjct 871 NGQCKNTPGSFRCVCDVGFH--LQDG-VCTDMDEC-VDPLQCP-GQQCINSPGSYKC 922
Score = 34.3 bits (77), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Query 56 DVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADA 93
++DEC + C + CVN DGS+ C C G+ AA A
Sbjct 965 NIDECVSDQV-CGAQSVCVNTDGSYRCDCAPGYRAAGA 1001
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 21/40 (52%), Gaps = 4/40 (10%)
Query 50 DGHACKDVDEC-AAGTAECHVSAQCVNVDGSFECTCMAGF 88
+G C D++EC G E + CVN GS+ C C GF
Sbjct 216 NGTQCVDINECLQPGFCE---NGNCVNTRGSYSCVCNEGF 252
Score = 27.7 bits (60), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDG 51
+ +C NT GSY C CN G+ +S G
Sbjct 234 NGNCVNTRGSYSCVCNEGFLLDASHG 259
> hsa:222663 SCUBE3, CEGF3, DKFZp686B09105, DKFZp686B1223, DKFZp686D20108,
FLJ34743; signal peptide, CUB domain, EGF-like
3
Length=993
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 33/66 (50%), Positives = 39/66 (59%), Gaps = 3/66 (4%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N G+Y CTC G+ A DGH C DVDECA G C S CVN+ GS+EC C GF
Sbjct 86 CVNIPGNYRCTCYDGFHLAH-DGHNCLDVDECAEGNGGCQQS--CVNMMGSYECHCREGF 142
Query 89 EAADAK 94
+D +
Sbjct 143 FLSDNQ 148
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 31/68 (45%), Positives = 36/68 (52%), Gaps = 6/68 (8%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDEC-AAGTAECHVSAQCVNVDGSFECT 83
+ A C NT SY C C GY + DG CKDVDEC A C CVN+ G++ CT
Sbjct 42 IDAICQNTPRSYKCICKSGY---TGDGKHCKDVDECEREDNAGC--VHDCVNIPGNYRCT 96
Query 84 CMAGFEAA 91
C GF A
Sbjct 97 CYDGFHLA 104
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 24/60 (40%), Positives = 35/60 (58%), Gaps = 4/60 (6%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT GS+ C+C GY+ ++ C+D+DEC+ H+ CVN GSF+C C G+
Sbjct 292 CRNTVGSFECSCKKGYKLLINE-RNCQDIDECSFDRTCDHI---CVNTPGSFQCLCHRGY 347
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 21/45 (46%), Positives = 27/45 (60%), Gaps = 1/45 (2%)
Query 53 ACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADAKTCK 97
A +DVDEC GT CH+ A C N S++C C +G+ D K CK
Sbjct 26 AAQDVDECVEGTDNCHIDAICQNTPRSYKCICKSGY-TGDGKHCK 69
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/59 (42%), Positives = 31/59 (52%), Gaps = 4/59 (6%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAG 87
C NT GS+ C C+ GY C DVDEC+ C C+N GS++CTC AG
Sbjct 332 CVNTPGSFQCLCHRGYLLYGIT--HCGDVDECSINRGGCRFG--CINTPGSYQCTCPAG 386
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 22/53 (41%), Positives = 28/53 (52%), Gaps = 5/53 (9%)
Query 38 CTCNPGYEPASSDGHACKDVDECAAGTAEC-HVSAQCVNVDGSFECTCMAGFE 89
CTC G+ D CKD+DEC C H+ C N GSFEC+C G++
Sbjct 260 CTCPVGFM-LQPDRKTCKDIDECRLNNGGCDHI---CRNTVGSFECSCKKGYK 308
> mmu:108075 Ltbp4, 2310046A13Rik, MGC175966; latent transforming
growth factor beta binding protein 4; K08023 latent transforming
growth factor beta binding protein
Length=1600
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 32/66 (48%), Positives = 37/66 (56%), Gaps = 3/66 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H C NT+GS+ CTC PGY P G +C DVDEC+ + S C N DGSFEC C
Sbjct 867 HGECLNTDGSFTCTCAPGYRPGPR-GASCLDVDECS--EEDLCQSGICTNTDGSFECICP 923
Query 86 AGFEAA 91
G A
Sbjct 924 PGHRAG 929
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 28/67 (41%), Positives = 37/67 (55%), Gaps = 1/67 (1%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
LH C NT+GS+ C+C+ GY S C D++EC G C +C+N DGSF CTC
Sbjct 823 LHGQCTNTKGSFHCSCSTGYRAPSGQPGPCADINECLEGDF-CFPHGECLNTDGSFTCTC 881
Query 85 MAGFEAA 91
G+
Sbjct 882 APGYRPG 888
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 34/71 (47%), Positives = 43/71 (60%), Gaps = 6/71 (8%)
Query 29 CANTEGSYVCT--CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C NT GSY CT C+PGY+P + G C+DVDEC + C A C N+ GSF+C C
Sbjct 998 CENTPGSYRCTPACDPGYQP--TPGGGCQDVDECR-NRSFCGAHAMCQNLPGSFQCVCDQ 1054
Query 87 GFEAA-DAKTC 96
G+E A D + C
Sbjct 1055 GYEGARDGRHC 1065
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/66 (42%), Positives = 36/66 (54%), Gaps = 2/66 (3%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
C NT GS++C C GY+ A+ G +C+DVDEC C C N+ GSF C C
Sbjct 619 RGRCENTPGSFLCVCPAGYQ-AAPHGASCQDVDECTQSPGLCGRGV-CENLPGSFRCVCP 676
Query 86 AGFEAA 91
AGF +
Sbjct 677 AGFRGS 682
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 32/70 (45%), Positives = 37/70 (52%), Gaps = 2/70 (2%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
HA C N GS+ C C+ GYE + DG C DV+EC C SA C NV+GSF C C
Sbjct 1038 HAMCQNLPGSFQCVCDQGYE-GARDGRHCVDVNECETLQGVCG-SALCENVEGSFLCVCP 1095
Query 86 AGFEAADAKT 95
E D T
Sbjct 1096 NSPEEFDPMT 1105
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 35/64 (54%), Gaps = 2/64 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C NT GS+ C C G+ + G C+DVDEC+ + C +C N +GSF+C C
Sbjct 701 GRCDNTAGSFHCACPAGFR-SRGPGAPCQDVDECSRSPSPC-AYGRCENTEGSFKCVCPT 758
Query 87 GFEA 90
GF+
Sbjct 759 GFQP 762
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/65 (41%), Positives = 33/65 (50%), Gaps = 2/65 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C NT GS+ C C G++ A C DVDEC C +C N GSF C C A
Sbjct 578 GRCENTPGSFRCVCGTGFQ-AGPRATECLDVDECRRVPPPCD-RGRCENTPGSFLCVCPA 635
Query 87 GFEAA 91
G++AA
Sbjct 636 GYQAA 640
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 29/69 (42%), Gaps = 2/69 (2%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C Y C C+PG+ G C D+DEC C +C N GSF C C
Sbjct 536 GRCVPRPSGYTCACDPGFR-LGPQGTRCIDIDECRRVPTPC-APGRCENTPGSFRCVCGT 593
Query 87 GFEAADAKT 95
GF+A T
Sbjct 594 GFQAGPRAT 602
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 43/108 (39%), Gaps = 40/108 (37%)
Query 26 HASCANTEGSYVCTCNPGYEPASSD----------------------------------G 51
+ C NTEGS+ C C G++P ++ G
Sbjct 742 YGRCENTEGSFKCVCPTGFQPNAAGSECEDVDECENRLACPGQECVNSPGSFQCRACPVG 801
Query 52 H-----ACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADAK 94
H C DVDEC++GT C + QC N GSF C+C G+ A +
Sbjct 802 HHLHRGRCTDVDECSSGTP-CGLHGQCTNTKGSFHCSCSTGYRAPSGQ 848
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 32/65 (49%), Gaps = 3/65 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDEC--AAGTAECHVSAQCVNVDGSFECT 83
H C + C C GY +C DV+EC A T+ V+A+CVN DGSF C
Sbjct 1521 HGRCVRVPEGFTCDCFDGYR-LDITRMSCVDVNECDEAEATSPLCVNARCVNTDGSFRCI 1579
Query 84 CMAGF 88
C GF
Sbjct 1580 CRPGF 1584
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/71 (40%), Positives = 37/71 (52%), Gaps = 5/71 (7%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECA-AGTAECHVSAQCVNVDGSFECT--CM 85
C NT+GS+ C C PG+ A D +C D+DEC G A C S +C N GS+ C C
Sbjct 911 CTNTDGSFECICPPGHR-AGPDLASCLDIDECRERGPALCG-SQRCENSPGSYRCVRDCD 968
Query 86 AGFEAADAKTC 96
G+ TC
Sbjct 969 PGYHPGPEGTC 979
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 28/63 (44%), Positives = 32/63 (50%), Gaps = 7/63 (11%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACK-DVDECAAGTAECHVSAQCVNVDGSFECTCMAG 87
C N GS+ C C PA G AC+ DVDECA C +C N GSF C C AG
Sbjct 664 CENLPGSFRCVC-----PAGFRGSACEEDVDECAQQPPPCG-PGRCDNTAGSFHCACPAG 717
Query 88 FEA 90
F +
Sbjct 718 FRS 720
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/74 (37%), Positives = 37/74 (50%), Gaps = 8/74 (10%)
Query 29 CANTEGSYVCT--CNPGYEPASSDGHACKDVDECAA-GTAECHVSAQCVNVDGSFECT-- 83
C N+ GSY C C+PGY P C D+DEC G+A C +C N GS+ CT
Sbjct 954 CENSPGSYRCVRDCDPGYHPGPEG--TCDDIDECREYGSAICGAQ-RCENTPGSYRCTPA 1010
Query 84 CMAGFEAADAKTCK 97
C G++ C+
Sbjct 1011 CDPGYQPTPGGGCQ 1024
Score = 35.0 bits (79), Expect = 0.064, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 1 LPMVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGH 52
L + CV V + + + A S ++A C NT+GS+ C C PG+ P H
Sbjct 1541 LDITRMSCVDVNECDEAEATSPLCVNARCVNTDGSFRCICRPGFAPTHQPHH 1592
Score = 33.5 bits (75), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 20/49 (40%), Positives = 25/49 (51%), Gaps = 5/49 (10%)
Query 40 CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C PG+E + +C DVDECA G H +C N G + C C GF
Sbjct 277 CPPGFERVNG---SCVDVDECATGGRCQH--GECANTRGGYTCVCPDGF 320
Score = 32.0 bits (71), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 27/56 (48%), Gaps = 2/56 (3%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
C NT Y C C+ G+ + C D DECA C +CVN GS+ CTC
Sbjct 1246 CVNTAPGYSCYCSNGFY-YHAHRLECVDNDECADEEPACE-GGRCVNTVGSYHCTC 1299
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 21/46 (45%), Gaps = 0/46 (0%)
Query 43 GYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
GY S D A +DVDEC + S CVN + C C GF
Sbjct 1216 GYLVPSGDLSARRDVDECQLFQDQVCKSGVCVNTAPGYSCYCSNGF 1261
> mmu:13645 Egf, AI790464; epidermal growth factor; K04357 epidermal
growth factor
Length=1217
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 32/73 (43%), Positives = 42/73 (57%), Gaps = 4/73 (5%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
S + C NTEG YVC C+ GYE DG +C D+DEC G C +A C N +G + CT
Sbjct 890 STSSRCINTEGGYVCRCSEGYE---GDGISCFDIDECQRGAHNCGENAACTNTEGGYNCT 946
Query 84 CMAGFEAADAKTC 96
C AG ++ +C
Sbjct 947 C-AGRPSSPGLSC 958
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 36/66 (54%), Gaps = 4/66 (6%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAEC-HVSAQCVNVDGSFECTC 84
HA C + + C C G+ + DG+ C D+DEC ++C S++C+N +G + C C
Sbjct 850 HARCVSDGETAECQCLKGF---ARDGNLCSDIDECVLARSDCPSTSSRCINTEGGYVCRC 906
Query 85 MAGFEA 90
G+E
Sbjct 907 SEGYEG 912
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 31/69 (44%), Gaps = 4/69 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C S+ C GY S D C+DV+ECA C + C N GS+ CTC GF
Sbjct 336 CERDPKSHSSACAEGYT-LSRDRKYCEDVNECATQNHGCTLG--CENTPGSYHCTCPTGF 392
Query 89 -EAADAKTC 96
D K C
Sbjct 393 VLLPDGKQC 401
Score = 35.8 bits (81), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 31/69 (44%), Gaps = 4/69 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAG- 87
C NT GSY CTC G+ DG C ++ C ++C S CV C C AG
Sbjct 377 CENTPGSYHCTCPTGFV-LLPDGKQCHELVSCPGNVSKC--SHGCVLTSDGPRCICPAGS 433
Query 88 FEAADAKTC 96
D KTC
Sbjct 434 VLGRDGKTC 442
> mmu:17064 Cd93, 6030404G09Rik, AA145088, AA4.1, AW555904, C1qr1,
C1qrp, Ly68; CD93 antigen; K06702 CDW93 antigen
Length=644
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 29/60 (48%), Positives = 38/60 (63%), Gaps = 2/60 (3%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT GS+ C C GY+P+ AC+DVDECAA + C + C+N DGSF C+C G+
Sbjct 355 CVNTLGSFHCECWVGYQPSGPKEEACEDVDECAAANSPC--AQGCINTDGSFYCSCKEGY 412
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/63 (41%), Positives = 35/63 (55%), Gaps = 1/63 (1%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT+GS+ C+C GY + D C+D+DEC+ + C N DGSF C C G+
Sbjct 397 CINTDGSFYCSCKEGYIVSGEDSTQCEDIDECSDARGN-PCDSLCFNTDGSFRCGCPPGW 455
Query 89 EAA 91
E A
Sbjct 456 ELA 458
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 31/60 (51%), Gaps = 5/60 (8%)
Query 35 SYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADAK 94
+Y C C GY+ SS H C D+DEC + + CVN GSF C C G++ + K
Sbjct 322 NYTCRCPSGYQLDSSQVH-CVDIDEC----QDSPCAQDCVNTLGSFHCECWVGYQPSGPK 376
> tgo:TGME49_069930 hypothetical protein
Length=2869
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 32/75 (42%), Positives = 42/75 (56%), Gaps = 4/75 (5%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAG-TAECHVSAQC 73
E A+ S+ + +GSY C C GYE DG C+D+DECAA T+ CH +A C
Sbjct 2465 ENQASQSAPATSTVWFTPKGSYRCRCKVGYE---GDGLNCRDIDECAAEETSPCHPAALC 2521
Query 74 VNVDGSFECTCMAGF 88
+N GSF C C A +
Sbjct 2522 INTKGSFRCVCKAHY 2536
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 32/100 (32%), Positives = 48/100 (48%), Gaps = 18/100 (18%)
Query 11 VPQGEASAAHSSTSLHASCANTEGSYVCT------------CNPGYEPASSDGHACKDVD 58
VPQ + +S S+ A C + EG C C+ GY ++D C+D+D
Sbjct 2545 VPQSDLVPGDTSNSV-AQCVDKEGRPSCLATLHCLPDGSCGCSEGYVRTTAD--TCEDID 2601
Query 59 ECAAGTAECHVS--AQCVNVDGSFECTCMAGFEAADAKTC 96
EC G A C + A C+N +G + C+C G+E D + C
Sbjct 2602 ECEEGLASCAPASLALCINTEGGYFCSCREGYE-GDGRQC 2640
> mmu:268935 Scube3, CEGF3, D030038I21Rik; signal peptide, CUB
domain, EGF-like 3
Length=993
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 33/66 (50%), Positives = 39/66 (59%), Gaps = 3/66 (4%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N G+Y CTC G+ A DGH C DVDECA G C S CVN+ GS+EC C GF
Sbjct 86 CVNIPGNYRCTCYDGFHLAH-DGHNCLDVDECAEGNGGCQQS--CVNMMGSYECHCRDGF 142
Query 89 EAADAK 94
+D +
Sbjct 143 FLSDNQ 148
Score = 50.8 bits (120), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 31/68 (45%), Positives = 36/68 (52%), Gaps = 6/68 (8%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDEC-AAGTAECHVSAQCVNVDGSFECT 83
+ A C NT SY C C GY + DG CKDVDEC A C CVN+ G++ CT
Sbjct 42 IDAICQNTPRSYKCICKSGY---TGDGKHCKDVDECEREDNAGC--VHDCVNIPGNYRCT 96
Query 84 CMAGFEAA 91
C GF A
Sbjct 97 CYDGFHLA 104
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/51 (45%), Positives = 30/51 (58%), Gaps = 1/51 (1%)
Query 47 ASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADAKTCK 97
A+ G A +DVDEC GT CH+ A C N S++C C +G+ D K CK
Sbjct 20 AAQHGKAAQDVDECVEGTDNCHIDAICQNTPRSYKCICKSGY-TGDGKHCK 69
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 24/60 (40%), Positives = 36/60 (60%), Gaps = 4/60 (6%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT GS+ C+C GY+ ++ +C+D+DEC+ H+ CVN GSF+C C G+
Sbjct 292 CRNTVGSFECSCKKGYKLLINE-RSCQDIDECSFDRTCDHM---CVNTPGSFQCLCHRGY 347
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/59 (42%), Positives = 31/59 (52%), Gaps = 4/59 (6%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAG 87
C NT GS+ C C+ GY C DVDEC+ C C+N GS++CTC AG
Sbjct 332 CVNTPGSFQCLCHRGYLLYGVT--HCGDVDECSINKGGCRFG--CINTPGSYQCTCPAG 386
Score = 37.0 bits (84), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 21/53 (39%), Positives = 28/53 (52%), Gaps = 5/53 (9%)
Query 38 CTCNPGYEPASSDGHACKDVDECAAGTAEC-HVSAQCVNVDGSFECTCMAGFE 89
C+C G+ D CKD+DEC C H+ C N GSFEC+C G++
Sbjct 260 CSCPVGFM-LQPDRKTCKDIDECRLNNGGCDHI---CRNTVGSFECSCKKGYK 308
> hsa:2192 FBLN1, FBLN, FIBL1; fibulin 1
Length=683
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 32/79 (40%), Positives = 47/79 (59%), Gaps = 2/79 (2%)
Query 21 SSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSF 80
SS+ CAN GSY C C GY+ + DG C+D+DECA T S +C+N+ GSF
Sbjct 446 SSSPCSQECANVYGSYQCYCRRGYQLSDVDGVTCEDIDECALPTGGHICSYRCINIPGSF 505
Query 81 ECTC-MAGFEAA-DAKTCK 97
+C+C +G+ A + + C+
Sbjct 506 QCSCPSSGYRLAPNGRNCQ 524
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 42/66 (63%), Gaps = 5/66 (7%)
Query 28 SCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAG 87
C NT GSY+C+C+ G+ S DG +C+D++EC++ S +C NV GS++C C G
Sbjct 414 KCENTLGSYLCSCSVGFR-LSVDGRSCEDINECSSSPC----SQECANVYGSYQCYCRRG 468
Query 88 FEAADA 93
++ +D
Sbjct 469 YQLSDV 474
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 41/73 (56%), Gaps = 5/73 (6%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFEC----TC 84
C +T VC+C GY+ SDG +C+DV+EC G+ C + C+N GSF C +C
Sbjct 190 CRDTGDEVVCSCFVGYQ-LLSDGVSCEDVNECITGSHSCRLGESCINTVGSFRCQRDSSC 248
Query 85 MAGFEAADAKTCK 97
G+E + +CK
Sbjct 249 GTGYELTEDNSCK 261
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 34/66 (51%), Gaps = 6/66 (9%)
Query 28 SCANTEGSYVC-----TCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFEC 82
+C NTEGSY C C GY + +G C DVDECA C +CVN GSF C
Sbjct 324 TCINTEGSYTCQKNVPNCGRGYH-LNEEGTRCVDVDECAPPAEPCGKGHRCVNSPGSFRC 382
Query 83 TCMAGF 88
C G+
Sbjct 383 ECKTGY 388
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 37/73 (50%), Gaps = 5/73 (6%)
Query 29 CANTEGSYVCTC-NPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT---C 84
C N GS+ C+C + GY A +G C+D+DEC G C ++ C N+ G F C C
Sbjct 498 CINIPGSFQCSCPSSGYRLAP-NGRNCQDIDECVTGIHNCSINETCFNIQGGFRCLAFEC 556
Query 85 MAGFEAADAKTCK 97
+ + A C+
Sbjct 557 PENYRRSAATRCE 569
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 30/75 (40%), Positives = 39/75 (52%), Gaps = 10/75 (13%)
Query 22 STSLHASCANTEGSYVC----TCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVD 77
S L SC NT GS+ C +C GYE ++ ++CKD+DEC +G C C N
Sbjct 226 SCRLGESCINTVGSFRCQRDSSCGTGYE--LTEDNSCKDIDECESGIHNCLPDFICQNTL 283
Query 78 GSFECT----CMAGF 88
GSF C C +GF
Sbjct 284 GSFRCRPKLQCKSGF 298
Score = 36.2 bits (82), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 35/72 (48%), Gaps = 7/72 (9%)
Query 29 CANTEGSYVCTCNPGYEPASSDG--HACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C N+ GS+ C C GY DG C DV+EC +C N GS+ C+C
Sbjct 373 CVNSPGSFRCECKTGY---YFDGISRMCVDVNECQRYPGR-LCGHKCENTLGSYLCSCSV 428
Query 87 GFE-AADAKTCK 97
GF + D ++C+
Sbjct 429 GFRLSVDGRSCE 440
Score = 32.7 bits (73), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 30/58 (51%), Gaps = 6/58 (10%)
Query 29 CANTEGSYVCT----CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFEC 82
C NT GS+ C C G+ + C D++EC + +A C + C+N +GS+ C
Sbjct 279 CQNTLGSFRCRPKLQCKSGFIQDALGN--CIDINECLSISAPCPIGHTCINTEGSYTC 334
> dre:569485 si:ch211-29n12.1
Length=801
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 30/75 (40%), Positives = 37/75 (49%), Gaps = 8/75 (10%)
Query 26 HASCANTEGSYVCTCNPGYEPAS-------SDGHACKDVDECAAGTAECHVSAQCVNVDG 78
H C NT GSY CTC GY P++ +DG C D+DEC C C N DG
Sbjct 103 HTKCHNTHGSYFCTCLAGYSPSNQMNTFIPNDGTYCHDIDECVV-QGVCGEGGLCQNTDG 161
Query 79 SFECTCMAGFEAADA 93
F C+C G++ D
Sbjct 162 DFTCSCQTGYKVQDG 176
Score = 35.8 bits (81), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 30/59 (50%), Gaps = 3/59 (5%)
Query 34 GSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAAD 92
G VC C G+ + C+D DEC G C +C N GS+ CTC+AG+ ++
Sbjct 70 GGKVCNCMYGF--VGNGRTYCQDKDECQLGKI-CGDHTKCHNTHGSYFCTCLAGYSPSN 125
> mmu:114249 Npnt, 1110009H02Rik, AA682063, AI314031, Nctn, POEM;
nephronectin; K06824 nephronectin
Length=561
Score = 58.5 bits (140), Expect = 4e-09, Method: Composition-based stats.
Identities = 31/86 (36%), Positives = 46/86 (53%), Gaps = 3/86 (3%)
Query 6 RQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDG-HACKDVDECAAGT 64
R CV + E + S C NT GSY+C C+ G++ G + C D+DEC+ G
Sbjct 165 RTCVDI--DECATGRVSCPRFRQCVNTFGSYICKCHTGFDLMYIGGKYQCHDIDECSLGQ 222
Query 65 AECHVSAQCVNVDGSFECTCMAGFEA 90
+C A+C N+ GS++C C G+E
Sbjct 223 HQCSSYARCYNIHGSYKCQCRDGYEG 248
Score = 51.2 bits (121), Expect = 7e-07, Method: Composition-based stats.
Identities = 32/84 (38%), Positives = 43/84 (51%), Gaps = 4/84 (4%)
Query 9 VFVPQGEASAAHSST--SLHASCANTEGSYVCTC-NPGYEPASSDGHACKDVDECAAGTA 65
+ +P G S+A S + + C +G C C +PG + A DG C D+DECA G
Sbjct 120 MLLPDGSCSSALSCSMANCQYGCDVVKGQVRCQCPSPGLQLAP-DGRTCVDIDECATGRV 178
Query 66 ECHVSAQCVNVDGSFECTCMAGFE 89
C QCVN GS+ C C GF+
Sbjct 179 SCPRFRQCVNTFGSYICKCHTGFD 202
Score = 32.0 bits (71), Expect = 0.51, Method: Composition-based stats.
Identities = 21/72 (29%), Positives = 33/72 (45%), Gaps = 11/72 (15%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHAC-KDVDECAAGTAECHVSAQCVNVDGSFECTC 84
H C G C C+PG+ G C +D++EC C +C+N GS++C C
Sbjct 66 HGECV---GPNKCKCHPGFA-----GKTCNQDLNECGLKPRPC--KHRCMNTFGSYKCYC 115
Query 85 MAGFEAADAKTC 96
+ G+ +C
Sbjct 116 LNGYMLLPDGSC 127
> hsa:7369 UMOD, ADMCKD2, FJHN, HNFJ, HNFJ1, MCKD2, THGP, THP;
uromodulin
Length=640
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 33/68 (48%), Positives = 40/68 (58%), Gaps = 3/68 (4%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECA-AGTAECHVSAQCVNVDGSFEC 82
S ++SC NT GS+ C C G+ S G C DVDECA G + CH A CVNV GS+ C
Sbjct 78 SANSSCVNTPGSFSCVCPEGFR--LSPGLGCTDVDECAEPGLSHCHALATCVNVVGSYLC 135
Query 83 TCMAGFEA 90
C AG+
Sbjct 136 VCPAGYRG 143
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 36/72 (50%), Gaps = 4/72 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAA-GTAECHVSAQCVNVDGSFECTC 84
+A+C E CTC G+ + DG C D+DECA G C ++ CVN GSF C C
Sbjct 38 NATCTEDEAVTTCTCQEGF---TGDGLTCVDLDECAIPGAHNCSANSSCVNTPGSFSCVC 94
Query 85 MAGFEAADAKTC 96
GF + C
Sbjct 95 PEGFRLSPGLGC 106
> cel:F54F3.1 nid-1; NIDogen (basement membrane protein family
member (nid-1); K06826 nidogen (entactin)
Length=1584
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 32/85 (37%), Positives = 43/85 (50%), Gaps = 5/85 (5%)
Query 16 ASAAHSSTSLHASCANTEGSYVCTCNPGYEPA----SSDGHACKDVDECAAGTAECHVSA 71
A H T + C + SY C C PGY+ A S G C+D+DEC G C A
Sbjct 664 APGRHQCTLPNMKCRVVDPSYRCECEPGYQAAHDASSHIGWICQDLDECQRGDHNCDQHA 723
Query 72 QCVNVDGSFECTCMAGFEAADAKTC 96
+C N GSF C C+ G++ D ++C
Sbjct 724 KCTNRPGSFSCQCLQGYQ-GDGRSC 747
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/76 (35%), Positives = 39/76 (51%), Gaps = 7/76 (9%)
Query 24 SLHASCAN--TEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVD-GSF 80
S+HA CA T G+Y C CN GY + +GH C + C + C +A CV + G +
Sbjct 1207 SIHAYCAQNPTSGAYQCKCNAGY---NGNGHLCVSMSSCLDDRSLCDENADCVPGEAGHY 1263
Query 81 ECTCMAGFEAADAKTC 96
C C G+ D ++C
Sbjct 1264 VCNCHYGYH-GDGRSC 1278
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 35/67 (52%), Gaps = 8/67 (11%)
Query 34 GSYVCTCNPGYEPASSDGHA-CKDVDEC-AAGTAECHVSAQCVN--VDGSFECTCMAGFE 89
G Y+C C PG+ S DG C+ D+C + + C+ +A CV + + C C+ GF+
Sbjct 1086 GEYICQCLPGF---SGDGFINCRGADQCNPSNPSACYQNAHCVYDAILNAHACKCVDGFK 1142
Query 90 AADAKTC 96
D +C
Sbjct 1143 -GDGTSC 1148
> mmu:14118 Fbn1, AI536462, B430209H23, Fib-1, Tsk; fibrillin
1; K06825 fibrillin 1
Length=2873
Score = 58.5 bits (140), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 33/72 (45%), Positives = 42/72 (58%), Gaps = 3/72 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ +C NT GS+ C CN G+ S + C DVDECA G + QCVN GSF+C C
Sbjct 1904 NGTCRNTIGSFNCRCNHGF--ILSHNNDCIDVDECATGNGNLCRNGQCVNTVGSFQCRCN 1961
Query 86 AGFEAA-DAKTC 96
G+E A D +TC
Sbjct 1962 EGYEVAPDGRTC 1973
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 45/73 (61%), Gaps = 4/73 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT+GS+ C CN G+ + DG C+D+DEC+ ++ C+N DGSF+C C
Sbjct 545 NGRCINTDGSFHCVCNAGFH-VTRDGKNCEDMDECSIRNM--CLNGMCINEDGSFKCICK 601
Query 86 AGFE-AADAKTCK 97
GF+ A+D + CK
Sbjct 602 PGFQLASDGRYCK 614
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 32/71 (45%), Positives = 40/71 (56%), Gaps = 4/71 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
HA C NT GS+ C+C+PG+ DG C D+DEC+ GT C A C N GS+ C C
Sbjct 1338 HAVCTNTAGSFKCSCSPGW---IGDGIKCTDLDECSNGTHMCSQHADCKNTMGSYRCLCK 1394
Query 86 AGFEAADAKTC 96
G+ D TC
Sbjct 1395 DGY-TGDGFTC 1404
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 35/84 (41%), Positives = 43/84 (51%), Gaps = 5/84 (5%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E S S HA C NT GSY C C GY + DG C D+DEC+ C + QC+
Sbjct 1368 ECSNGTHMCSQHADCKNTMGSYRCLCKDGY---TGDGFTCTDLDECSENLNLCG-NGQCL 1423
Query 75 NVDGSFECTCMAGF-EAADAKTCK 97
N G + C C GF +AD K C+
Sbjct 1424 NAPGGYRCECDMGFVPSADGKACE 1447
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 42/73 (57%), Gaps = 4/73 (5%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
L+ C N +GS+ C C PG++ A SDG CKD++EC T ++ +CVN DGS+ C C
Sbjct 585 LNGMCINEDGSFKCICKPGFQLA-SDGRYCKDINECE--TPGICMNGRCVNTDGSYRCEC 641
Query 85 MAGFEAA-DAKTC 96
G D + C
Sbjct 642 FPGLAVGLDGRVC 654
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/90 (38%), Positives = 46/90 (51%), Gaps = 7/90 (7%)
Query 8 CVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAEC 67
C+ + + E SA + H C N G Y C CNPGY P + D C D+DEC+ C
Sbjct 1155 CIDINECELSA---NLCPHGRCVNLIGKYQCACNPGYHP-THDRLFCVDIDECSIMNGGC 1210
Query 68 HVSAQCVNVDGSFECTCMAGFE-AADAKTC 96
C N DGS+EC+C GF D ++C
Sbjct 1211 ETF--CTNSDGSYECSCQPGFALMPDQRSC 1238
Score = 55.8 bits (133), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 41/80 (51%), Gaps = 2/80 (2%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E + + + + C NT GS+ C CN GYE A DG C D++EC +C C
Sbjct 1935 ECATGNGNLCRNGQCVNTVGSFQCRCNEGYEVAP-DGRTCVDINECVLDPGKC-APGTCQ 1992
Query 75 NVDGSFECTCMAGFEAADAK 94
N+DGS+ C C G+ + K
Sbjct 1993 NLDGSYRCICPPGYSLQNDK 2012
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 40/72 (55%), Gaps = 3/72 (4%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C N +GSY C C GY+ ++ C+D+DEC C+ + +C+N DGSF C C A
Sbjct 504 GECINNQGSYTCHCRAGYQ-STLTRTECRDIDECLQNGRICN-NGRCINTDGSFHCVCNA 561
Query 87 GFEAA-DAKTCK 97
GF D K C+
Sbjct 562 GFHVTRDGKNCE 573
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 38/72 (52%), Gaps = 5/72 (6%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H C NT+GSY C C GY +G+ C D DEC+ G + C NV G FECTC
Sbjct 2141 HGQCINTDGSYRCECPFGY---ILEGNECVDTDECSVGNP--CGNGTCKNVIGGFECTCE 2195
Query 86 AGFEAADAKTCK 97
GFE TC+
Sbjct 2196 EGFEPGPMMTCE 2207
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/60 (50%), Positives = 33/60 (55%), Gaps = 3/60 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NTEGSY C+C GY DG +CKD+DECA C CVN G F C C GF
Sbjct 2461 CKNTEGSYQCSCPKGY-ILQEDGRSCKDLDECATKQHNCQF--LCVNTIGGFTCKCPPGF 2517
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 29/69 (42%), Positives = 38/69 (55%), Gaps = 3/69 (4%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N++GSY C+C PG+ D +C D+DEC C QC N+ G + C C GF
Sbjct 1214 CTNSDGSYECSCQPGFA-LMPDQRSCTDIDECEDNPNICD-GGQCTNIPGEYRCLCYDGF 1271
Query 89 EAA-DAKTC 96
A+ D KTC
Sbjct 1272 MASEDMKTC 1280
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 38/79 (48%), Gaps = 2/79 (2%)
Query 16 ASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVN 75
A H T C N G+Y+C C PGY+ DG C D +EC C + +C+N
Sbjct 2254 AEGKHDCTEKQMECKNLIGTYMCICGPGYQ-RRPDGEGCIDENECQTKPGICE-NGRCLN 2311
Query 76 VDGSFECTCMAGFEAADAK 94
GS+ C C GF A+ +
Sbjct 2312 TLGSYTCECNDGFTASPTQ 2330
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 31/73 (42%), Positives = 38/73 (52%), Gaps = 3/73 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N G+Y C CN GYE G C D++EC + C + QC N GSF CTC
Sbjct 738 NGICENLRGTYKCICNSGYE-VDITGKNCVDINECVLNSLLCD-NGQCRNTPGSFVCTCP 795
Query 86 AGF-EAADAKTCK 97
GF D KTC+
Sbjct 796 KGFVYKPDLKTCE 808
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 36/70 (51%), Gaps = 1/70 (1%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
L +C NT+GS++C C+ GY C D++EC G C A C N GSF+C+C
Sbjct 1294 LSGTCENTKGSFICHCDMGYS-GKKGKTGCTDINECEIGAHNCGRHAVCTNTAGSFKCSC 1352
Query 85 MAGFEAADAK 94
G+ K
Sbjct 1353 SPGWIGDGIK 1362
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 35/70 (50%), Gaps = 2/70 (2%)
Query 22 STSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFE 81
S H C NT GS+ C C+ G+ S+ C D+DEC C QCVN G FE
Sbjct 1039 SLCTHGKCRNTIGSFKCRCDSGFA-LDSEERNCTDIDECRISPDLCG-RGQCVNTPGDFE 1096
Query 82 CTCMAGFEAA 91
C C G+E+
Sbjct 1097 CKCDEGYESG 1106
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 39/73 (53%), Gaps = 5/73 (6%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ +C N G + CTC G+EP C+D++ECA C + +CVN GS+EC C
Sbjct 2180 NGTCKNVIGGFECTCEEGFEPGPM--MTCEDINECAQNPLLC--AFRCVNTYGSYECKCP 2235
Query 86 AGFEAA-DAKTCK 97
G+ D + CK
Sbjct 2236 VGYVLREDRRMCK 2248
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 26/62 (41%), Positives = 33/62 (53%), Gaps = 4/62 (6%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
+C N +GSY C C PGY S C+D+DEC C + C N +GSF+C C
Sbjct 1989 GTCQNLDGSYRCICPPGY---SLQNDKCEDIDECVEEPEICALGT-CSNTEGSFKCLCPE 2044
Query 87 GF 88
GF
Sbjct 2045 GF 2046
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 39/73 (53%), Gaps = 4/73 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N GSY C C GY P + G AC D++EC C+ C N +GS++C+C
Sbjct 2417 NGECVNDRGSYHCICKTGYTPDIT-GTACVDLNECNQAPKPCNFI--CKNTEGSYQCSCP 2473
Query 86 AGF-EAADAKTCK 97
G+ D ++CK
Sbjct 2474 KGYILQEDGRSCK 2486
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 35/64 (54%), Gaps = 3/64 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N G Y C C+ G+ P S+DG AC+D+DEC+ V C N+ G F C C
Sbjct 1419 NGQCLNAPGGYRCECDMGFVP-SADGKACEDIDECSLPNI--CVFGTCHNLPGLFRCECE 1475
Query 86 AGFE 89
G+E
Sbjct 1476 IGYE 1479
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 35/70 (50%), Gaps = 3/70 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+A C NT GSY C C PGY S+ C D +EC C QC++ GSF C C
Sbjct 1823 NAECINTAGSYRCDCKPGYRLTSTG--QCNDRNECQEIPNICS-HGQCIDTVGSFYCLCH 1879
Query 86 AGFEAADAKT 95
GF+ +T
Sbjct 1880 TGFKTNVDQT 1889
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 26/62 (41%), Positives = 33/62 (53%), Gaps = 2/62 (3%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAEC-HVSAQCVNVDGSFECTCMAG 87
C NT GSY C C GY D CKD DECA G +C +C N+ G++ C C G
Sbjct 2223 CVNTYGSYECKCPVGYV-LREDRRMCKDEDECAEGKHDCTEKQMECKNLIGTYMCICGPG 2281
Query 88 FE 89
++
Sbjct 2282 YQ 2283
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 40/93 (43%), Gaps = 10/93 (10%)
Query 10 FVPQGEASAAHS-------STSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAA 62
FVP + A + + +C N G + C C GYE S G+ C DV+EC
Sbjct 1437 FVPSADGKACEDIDECSLPNICVFGTCHNLPGLFRCECEIGYELDRSGGN-CTDVNECLD 1495
Query 63 GTAECHVSAQCVNVDGSFECTCMAGFEAADAKT 95
T +S CVN GS+ C C FE +
Sbjct 1496 PTT--CISGNCVNTPGSYTCDCPPDFELNPTRV 1526
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/69 (39%), Positives = 31/69 (44%), Gaps = 2/69 (2%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C N G Y C C G+ AS D C DV+EC C +S C N GSF C C
Sbjct 1254 GQCTNIPGEYRCLCYDGFM-ASEDMKTCVDVNECDLNPNIC-LSGTCENTKGSFICHCDM 1311
Query 87 GFEAADAKT 95
G+ KT
Sbjct 1312 GYSGKKGKT 1320
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 31/84 (36%), Positives = 43/84 (51%), Gaps = 7/84 (8%)
Query 5 FRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGT 64
+ QCV + E +AH ASC NT GSY C C G++ G C+D++EC +
Sbjct 2604 WNQCV--DENECLSAHVCGG--ASCHNTLGSYKCMCPTGFQYEQFSG-GCQDINECGSSQ 2658
Query 65 AECHVSAQCVNVDGSFECTCMAGF 88
A C S C N +G + C C G+
Sbjct 2659 APC--SYGCSNTEGGYLCGCPPGY 2680
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 41/97 (42%), Gaps = 11/97 (11%)
Query 1 LPMVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDEC 60
LP+ +C +P + C N GS+ C C G+ + C+D+DEC
Sbjct 1765 LPVDIDECREIP---------GVCENGVCINMVGSFRCECPVGFF-YNDKLLVCEDIDEC 1814
Query 61 AAGTAECHVSAQCVNVDGSFECTCMAGFEAADAKTCK 97
G C +A+C+N GS+ C C G+ C
Sbjct 1815 QNGPV-CQRNAECINTAGSYRCDCKPGYRLTSTGQCN 1850
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 34/70 (48%), Gaps = 5/70 (7%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C T GSY C CN G++ C DVDEC +C+N GS+ C C
Sbjct 464 NGRCIPTPGSYRCECNKGFQLDIRG--ECIDVDECEKNPC---TGGECINNQGSYTCHCR 518
Query 86 AGFEAADAKT 95
AG+++ +T
Sbjct 519 AGYQSTLTRT 528
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 29/66 (43%), Gaps = 1/66 (1%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
C NT G + C C+ GYE C D+DEC C C N +GS+ C C
Sbjct 1085 RGQCVNTPGDFECKCDEGYESGFMMMKNCMDIDECQRDPLLCR-GGICHNTEGSYRCECP 1143
Query 86 AGFEAA 91
G + +
Sbjct 1144 PGHQLS 1149
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 28/60 (46%), Gaps = 3/60 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT G + C C PG+ + AC D +EC + C C N GSF C C GF
Sbjct 2502 CVNTIGGFTCKCPPGFTQHHT---ACIDNNECTSDINLCGSKGVCQNTPGSFTCECQRGF 2558
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 33/61 (54%), Gaps = 8/61 (13%)
Query 26 HASCANTEGSYVCTCNPG--YEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
+ C NT GS+VCTC G Y+P D C+D+DEC + ++ C N GSF C
Sbjct 780 NGQCRNTPGSFVCTCPKGFVYKP---DLKTCEDIDECESSPC---INGVCKNSPGSFICE 833
Query 84 C 84
C
Sbjct 834 C 834
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 24/60 (40%), Positives = 32/60 (53%), Gaps = 4/60 (6%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT GS+ C C G+ S G +C+DVDEC G C Q N+ G + C+C G+
Sbjct 2543 CQNTPGSFTCECQRGFSLDQS-GASCEDVDEC-EGNHRCQHGCQ--NIIGGYRCSCPQGY 2598
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 23/48 (47%), Positives = 26/48 (54%), Gaps = 2/48 (4%)
Query 40 CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAG 87
C G+ P G AC+DVDEC A C C+N GSFEC C AG
Sbjct 231 CRRGFIPNIRTG-ACQDVDECQAIPGMCQ-GGNCINTVGSFECKCPAG 276
Score = 34.7 bits (78), Expect = 0.072, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 31/62 (50%), Gaps = 2/62 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
+C NT GS+ C C G++ + C+D+DEC+ C +C N S+ C C
Sbjct 260 GNCINTVGSFECKCPAGHK-FNEVSQKCEDIDECSTIPGVCD-GGECTNTVSSYFCKCPP 317
Query 87 GF 88
GF
Sbjct 318 GF 319
Score = 31.6 bits (70), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 30/63 (47%), Gaps = 7/63 (11%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHV--SAQCVNVDGSFECTCMA 86
C N G Y C+C GY + C D +EC + HV A C N GS++C C
Sbjct 2583 CQNIIGGYRCSCPQGYL-QHYQWNQCVDENECLSA----HVCGGASCHNTLGSYKCMCPT 2637
Query 87 GFE 89
GF+
Sbjct 2638 GFQ 2640
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 22/43 (51%), Gaps = 2/43 (4%)
Query 55 KDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEA-ADAKTC 96
+D+DEC C +C+N GSF+C C G+ D + C
Sbjct 1607 EDIDECQELPGLCQ-GGKCINTFGSFQCRCPTGYYLNEDTRVC 1648
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 20/31 (64%), Gaps = 1/31 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDV 57
+C+NTEGS+ C C G+ SS G C+D+
Sbjct 2029 GTCSNTEGSFKCLCPEGFS-LSSTGRRCQDL 2058
Score = 28.9 bits (63), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 20/38 (52%), Gaps = 1/38 (2%)
Query 51 GHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
G C+D++EC C + CVN GSF+C C G
Sbjct 907 GTQCEDINECEVFPGVCK-NGLCVNSRGSFKCECPNGM 943
Score = 28.1 bits (61), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 26/55 (47%), Gaps = 10/55 (18%)
Query 1 LPMVFR-QCVFVPQGEASAAH---------SSTSLHASCANTEGSYVCTCNPGYE 45
LP +FR +C + + S + +T + +C NT GSY C C P +E
Sbjct 1466 LPGLFRCECEIGYELDRSGGNCTDVNECLDPTTCISGNCVNTPGSYTCDCPPDFE 1520
Score = 27.7 bits (60), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 16/31 (51%), Gaps = 1/31 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDV 57
C NT SY C C PG+ S DG C DV
Sbjct 302 GECTNTVSSYFCKCPPGFY-TSPDGTRCVDV 331
> hsa:2200 FBN1, FBN, MASS, MFS1, OCTD, SGS, SSKS, WMS; fibrillin
1; K06825 fibrillin 1
Length=2871
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 43/72 (59%), Gaps = 3/72 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ +C NT GS+ C CN G+ S + C DVDECA+G + QC+N GSF+C C
Sbjct 1902 NGTCRNTIGSFNCRCNHGF--ILSHNNDCIDVDECASGNGNLCRNGQCINTVGSFQCQCN 1959
Query 86 AGFEAA-DAKTC 96
G+E A D +TC
Sbjct 1960 EGYEVAPDGRTC 1971
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 45/73 (61%), Gaps = 4/73 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT+GS+ C CN G+ + DG C+D+DEC+ ++ C+N DGSF+C C
Sbjct 543 NGRCINTDGSFHCVCNAGFH-VTRDGKNCEDMDECSIRNM--CLNGMCINEDGSFKCICK 599
Query 86 AGFE-AADAKTCK 97
GF+ A+D + CK
Sbjct 600 PGFQLASDGRYCK 612
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 32/71 (45%), Positives = 40/71 (56%), Gaps = 4/71 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
HA C NT GS+ C+C+PG+ DG C D+DEC+ GT C A C N GS+ C C
Sbjct 1336 HAVCTNTAGSFKCSCSPGW---IGDGIKCTDLDECSNGTHMCSQHADCKNTMGSYRCLCK 1392
Query 86 AGFEAADAKTC 96
G+ D TC
Sbjct 1393 EGY-TGDGFTC 1402
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 35/84 (41%), Positives = 43/84 (51%), Gaps = 5/84 (5%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E S S HA C NT GSY C C GY + DG C D+DEC+ C + QC+
Sbjct 1366 ECSNGTHMCSQHADCKNTMGSYRCLCKEGY---TGDGFTCTDLDECSENLNLCG-NGQCL 1421
Query 75 NVDGSFECTCMAGF-EAADAKTCK 97
N G + C C GF +AD K C+
Sbjct 1422 NAPGGYRCECDMGFVPSADGKACE 1445
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 42/73 (57%), Gaps = 4/73 (5%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
L+ C N +GS+ C C PG++ A SDG CKD++EC T ++ +CVN DGS+ C C
Sbjct 583 LNGMCINEDGSFKCICKPGFQLA-SDGRYCKDINECE--TPGICMNGRCVNTDGSYRCEC 639
Query 85 MAGFEAA-DAKTC 96
G D + C
Sbjct 640 FPGLAVGLDGRVC 652
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 40/72 (55%), Gaps = 3/72 (4%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C N +GSY C C GY+ ++ C+D+DEC C+ + +C+N DGSF C C A
Sbjct 502 GECINNQGSYTCQCRAGYQ-STLTRTECRDIDECLQNGRICN-NGRCINTDGSFHCVCNA 559
Query 87 GFEAA-DAKTCK 97
GF D K C+
Sbjct 560 GFHVTRDGKNCE 571
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 42/80 (52%), Gaps = 2/80 (2%)
Query 15 EASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCV 74
E ++ + + + C NT GS+ C CN GYE A DG C D++EC +C C
Sbjct 1933 ECASGNGNLCRNGQCINTVGSFQCQCNEGYEVAP-DGRTCVDINECLLEPRKC-APGTCQ 1990
Query 75 NVDGSFECTCMAGFEAADAK 94
N+DGS+ C C G+ + K
Sbjct 1991 NLDGSYRCICPPGYSLQNEK 2010
Score = 54.7 bits (130), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 33/72 (45%), Positives = 38/72 (52%), Gaps = 5/72 (6%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
H C NT+GSY C C GY A G+ C D DEC+ G + C NV G FECTC
Sbjct 2139 HGQCINTDGSYRCECPFGYILA---GNECVDTDECSVGNP--CGNGTCKNVIGGFECTCE 2193
Query 86 AGFEAADAKTCK 97
GFE TC+
Sbjct 2194 EGFEPGPMMTCE 2205
Score = 54.3 bits (129), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 32/73 (43%), Positives = 39/73 (53%), Gaps = 3/73 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N G+Y C CN GYE S G C D++EC + C + QC N GSF CTC
Sbjct 736 NGICENLRGTYKCICNSGYE-VDSTGKNCVDINECVLNSLLCD-NGQCRNTPGSFVCTCP 793
Query 86 AGF-EAADAKTCK 97
GF D KTC+
Sbjct 794 KGFIYKPDLKTCE 806
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 30/69 (43%), Positives = 38/69 (55%), Gaps = 3/69 (4%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N+EGSY C+C PG+ D +C D+DEC C QC N+ G + C C GF
Sbjct 1212 CTNSEGSYECSCQPGFA-LMPDQRSCTDIDECEDNPNICD-GGQCTNIPGEYRCLCYDGF 1269
Query 89 EAA-DAKTC 96
A+ D KTC
Sbjct 1270 MASEDMKTC 1278
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/60 (50%), Positives = 33/60 (55%), Gaps = 3/60 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NTEGSY C+C GY DG +CKD+DECA C CVN G F C C GF
Sbjct 2459 CKNTEGSYQCSCPKGY-ILQEDGRSCKDLDECATKQHNCQF--LCVNTIGGFTCKCPPGF 2515
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 36/74 (48%), Gaps = 2/74 (2%)
Query 18 AAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVD 77
H T C N G+Y+C C PGY+ DG C D +EC C + +C+N
Sbjct 2254 GKHDCTEKQMECKNLIGTYMCICGPGYQ-RRPDGEGCVDENECQTKPGICE-NGRCLNTR 2311
Query 78 GSFECTCMAGFEAA 91
GS+ C C GF A+
Sbjct 2312 GSYTCECNDGFTAS 2325
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 36/70 (51%), Gaps = 1/70 (1%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
L +C NT+GS++C C+ GY C D++EC G C A C N GSF+C+C
Sbjct 1292 LSGTCENTKGSFICHCDMGYS-GKKGKTGCTDINECEIGAHNCGKHAVCTNTAGSFKCSC 1350
Query 85 MAGFEAADAK 94
G+ K
Sbjct 1351 SPGWIGDGIK 1360
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 38/72 (52%), Gaps = 4/72 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N G Y C CNPGY ++ D C D+DEC+ C C N +GS+EC+C
Sbjct 1168 NGRCVNLIGKYQCACNPGYH-STPDRLFCVDIDECSIMNGGCETF--CTNSEGSYECSCQ 1224
Query 86 AGFE-AADAKTC 96
GF D ++C
Sbjct 1225 PGFALMPDQRSC 1236
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 35/70 (50%), Gaps = 2/70 (2%)
Query 22 STSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFE 81
S H C NT GS+ C C+ G+ S+ C D+DEC C QCVN G FE
Sbjct 1037 SLCTHGKCRNTIGSFKCRCDSGFA-LDSEERNCTDIDECRISPDLCG-RGQCVNTPGDFE 1094
Query 82 CTCMAGFEAA 91
C C G+E+
Sbjct 1095 CKCDEGYESG 1104
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 29/70 (41%), Positives = 36/70 (51%), Gaps = 3/70 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+A C NT GSY C C PGY S+ C D +EC C QC++ GSF C C
Sbjct 1821 NAECINTAGSYRCDCKPGYRFTSTG--QCNDRNECQEIPNICS-HGQCIDTVGSFYCLCH 1877
Query 86 AGFEAADAKT 95
GF+ D +T
Sbjct 1878 TGFKTNDDQT 1887
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 26/62 (41%), Positives = 33/62 (53%), Gaps = 4/62 (6%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
+C N +GSY C C PGY S C+D+DEC C + C N +GSF+C C
Sbjct 1987 GTCQNLDGSYRCICPPGY---SLQNEKCEDIDECVEEPEICALGT-CSNTEGSFKCLCPE 2042
Query 87 GF 88
GF
Sbjct 2043 GF 2044
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 39/73 (53%), Gaps = 5/73 (6%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ +C N G + CTC G+EP C+D++ECA C + +CVN GS+EC C
Sbjct 2178 NGTCKNVIGGFECTCEEGFEPGPM--MTCEDINECAQNPLLC--AFRCVNTYGSYECKCP 2233
Query 86 AGFEAA-DAKTCK 97
G+ D + CK
Sbjct 2234 VGYVLREDRRMCK 2246
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 35/64 (54%), Gaps = 3/64 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N G Y C C+ G+ P S+DG AC+D+DEC+ V C N+ G F C C
Sbjct 1417 NGQCLNAPGGYRCECDMGFVP-SADGKACEDIDECSLPNI--CVFGTCHNLPGLFRCECE 1473
Query 86 AGFE 89
G+E
Sbjct 1474 IGYE 1477
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 39/73 (53%), Gaps = 4/73 (5%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C N GSY C C GY P + G +C D++EC C+ C N +GS++C+C
Sbjct 2415 NGECVNDRGSYHCICKTGYTPDIT-GTSCVDLNECNQAPKPCNFI--CKNTEGSYQCSCP 2471
Query 86 AGF-EAADAKTCK 97
G+ D ++CK
Sbjct 2472 KGYILQEDGRSCK 2484
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAEC-HVSAQCVNVDGSFECTCMAG 87
C NT GSY C C GY D CKD DEC G +C +C N+ G++ C C G
Sbjct 2221 CVNTYGSYECKCPVGYV-LREDRRMCKDEDECEEGKHDCTEKQMECKNLIGTYMCICGPG 2279
Query 88 FE 89
++
Sbjct 2280 YQ 2281
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 40/93 (43%), Gaps = 10/93 (10%)
Query 10 FVPQGEASAAHS-------STSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAA 62
FVP + A + + +C N G + C C GYE S G+ C DV+EC
Sbjct 1435 FVPSADGKACEDIDECSLPNICVFGTCHNLPGLFRCECEIGYELDRSGGN-CTDVNECLD 1493
Query 63 GTAECHVSAQCVNVDGSFECTCMAGFEAADAKT 95
T +S CVN GS+ C C FE +
Sbjct 1494 PTT--CISGNCVNTPGSYICDCPPDFELNPTRV 1524
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/69 (39%), Positives = 31/69 (44%), Gaps = 2/69 (2%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C N G Y C C G+ AS D C DV+EC C +S C N GSF C C
Sbjct 1252 GQCTNIPGEYRCLCYDGFM-ASEDMKTCVDVNECDLNPNIC-LSGTCENTKGSFICHCDM 1309
Query 87 GFEAADAKT 95
G+ KT
Sbjct 1310 GYSGKKGKT 1318
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 31/84 (36%), Positives = 43/84 (51%), Gaps = 7/84 (8%)
Query 5 FRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGT 64
+ QCV + E +AH ASC NT GSY C C G++ G C+D++EC +
Sbjct 2602 WNQCV--DENECLSAHICGG--ASCHNTLGSYKCMCPAGFQYEQFSG-GCQDINECGSAQ 2656
Query 65 AECHVSAQCVNVDGSFECTCMAGF 88
A C S C N +G + C C G+
Sbjct 2657 APC--SYGCSNTEGGYLCGCPPGY 2678
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 34/70 (48%), Gaps = 5/70 (7%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C T GSY C CN G++ C DVDEC +C+N GS+ C C
Sbjct 462 NGRCIPTPGSYRCECNKGFQLDLRG--ECIDVDECEKNPC---AGGECINNQGSYTCQCR 516
Query 86 AGFEAADAKT 95
AG+++ +T
Sbjct 517 AGYQSTLTRT 526
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 41/97 (42%), Gaps = 11/97 (11%)
Query 1 LPMVFRQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDEC 60
LP+ +C +P + C N GS+ C C G+ + C+D+DEC
Sbjct 1763 LPVDIDECREIP---------GVCENGVCINMVGSFRCECPVGFF-YNDKLLVCEDIDEC 1812
Query 61 AAGTAECHVSAQCVNVDGSFECTCMAGFEAADAKTCK 97
G C +A+C+N GS+ C C G+ C
Sbjct 1813 QNGPV-CQRNAECINTAGSYRCDCKPGYRFTSTGQCN 1848
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 29/66 (43%), Gaps = 1/66 (1%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
C NT G + C C+ GYE C D+DEC C C N +GS+ C C
Sbjct 1083 RGQCVNTPGDFECKCDEGYESGFMMMKNCMDIDECQRDPLLCR-GGVCHNTEGSYRCECP 1141
Query 86 AGFEAA 91
G + +
Sbjct 1142 PGHQLS 1147
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 28/60 (46%), Gaps = 3/60 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT G + C C PG+ + +C D +EC + C C N GSF C C GF
Sbjct 2500 CVNTIGGFTCKCPPGFTQHHT---SCIDNNECTSDINLCGSKGICQNTPGSFTCECQRGF 2556
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 33/61 (54%), Gaps = 8/61 (13%)
Query 26 HASCANTEGSYVCTCNPG--YEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
+ C NT GS+VCTC G Y+P D C+D+DEC + ++ C N GSF C
Sbjct 778 NGQCRNTPGSFVCTCPKGFIYKP---DLKTCEDIDECESSPC---INGVCKNSPGSFICE 831
Query 84 C 84
C
Sbjct 832 C 832
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 31/60 (51%), Gaps = 4/60 (6%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT GS+ C C G+ G +C+DVDEC G C Q N+ G + C+C G+
Sbjct 2541 CQNTPGSFTCECQRGFS-LDQTGSSCEDVDEC-EGNHRCQHGCQ--NIIGGYRCSCPQGY 2596
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 28/54 (51%), Gaps = 2/54 (3%)
Query 40 CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADA 93
C G+ P G AC+DVDEC A C C+N GSFEC C AG + +
Sbjct 231 CRRGFIPNIRTG-ACQDVDECQAIPGLCQ-GGNCINTVGSFECKCPAGHKLNEV 282
Score = 35.4 bits (80), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 31/62 (50%), Gaps = 2/62 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
+C NT GS+ C C G++ + C+D+DEC+ C +C N S+ C C
Sbjct 260 GNCINTVGSFECKCPAGHK-LNEVSQKCEDIDECSTIPGICE-GGECTNTVSSYFCKCPP 317
Query 87 GF 88
GF
Sbjct 318 GF 319
Score = 33.1 bits (74), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 25/49 (51%), Gaps = 3/49 (6%)
Query 40 CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C GY + G C+D+DEC C + CVN GSF+C C +G
Sbjct 896 CGKGY--SRIKGTQCEDIDECEVFPGVCK-NGLCVNTRGSFKCQCPSGM 941
Score = 33.1 bits (74), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 31/65 (47%), Gaps = 11/65 (16%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAA----GTAECHVSAQCVNVDGSFECTC 84
C N G Y C+C GY + C D +EC + G A CH N GS++C C
Sbjct 2581 CQNIIGGYRCSCPQGYL-QHYQWNQCVDENECLSAHICGGASCH------NTLGSYKCMC 2633
Query 85 MAGFE 89
AGF+
Sbjct 2634 PAGFQ 2638
Score = 31.2 bits (69), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 28/67 (41%), Gaps = 3/67 (4%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C NT GS+ C C GY + D C DV+EC T C N G++ C C
Sbjct 1620 GKCINTFGSFQCRCPTGYY-LNEDTRVCDDVNECE--TPGICGPGTCYNTVGNYTCICPP 1676
Query 87 GFEAADA 93
+ +
Sbjct 1677 DYMQVNG 1683
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 22/43 (51%), Gaps = 2/43 (4%)
Query 55 KDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEA-ADAKTC 96
+D+DEC C +C+N GSF+C C G+ D + C
Sbjct 1605 EDIDECQELPGLCQ-GGKCINTFGSFQCRCPTGYYLNEDTRVC 1646
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 27/55 (49%), Gaps = 10/55 (18%)
Query 1 LPMVFR-QCVFVPQGEASAAH---------SSTSLHASCANTEGSYVCTCNPGYE 45
LP +FR +C + + S + +T + +C NT GSY+C C P +E
Sbjct 1464 LPGLFRCECEIGYELDRSGGNCTDVNECLDPTTCISGNCVNTPGSYICDCPPDFE 1518
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 20/31 (64%), Gaps = 1/31 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDV 57
+C+NTEGS+ C C G+ SS G C+D+
Sbjct 2027 GTCSNTEGSFKCLCPEGFS-LSSSGRRCQDL 2056
Score = 28.1 bits (61), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 16/31 (51%), Gaps = 1/31 (3%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDV 57
C NT SY C C PG+ S DG C DV
Sbjct 302 GECTNTVSSYFCKCPPGFY-TSPDGTRCIDV 331
> mmu:16998 Ltbp3, Ltbp2, mFLJ00070; latent transforming growth
factor beta binding protein 3; K08023 latent transforming
growth factor beta binding protein
Length=1253
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 32/67 (47%), Positives = 40/67 (59%), Gaps = 3/67 (4%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N GSY CTC GYEPA DG +C DVDEC AG C C N GSF+C C++G+
Sbjct 756 CENLPGSYRCTCAQGYEPA-QDGLSCIDVDECEAGKV-CQ-DGICTNTPGSFQCQCLSGY 812
Query 89 EAADAKT 95
+ ++
Sbjct 813 HLSRDRS 819
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 31/71 (43%), Positives = 38/71 (53%), Gaps = 5/71 (7%)
Query 22 STSLHASCANTEGSYVCT-CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSF 80
ST C N GS+ C C PGY S G AC+DV+EC+ GT C N+ GS+
Sbjct 708 STCPDGKCENKPGSFKCIACQPGYR--SQGGGACRDVNECSEGTP--CSPGWCENLPGSY 763
Query 81 ECTCMAGFEAA 91
CTC G+E A
Sbjct 764 RCTCAQGYEPA 774
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 35/69 (50%), Gaps = 3/69 (4%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT GS+ C C GY S D C+D+DEC A C + C+N +GS+ C C G
Sbjct 797 CTNTPGSFQCQCLSGYH-LSRDRSRCEDIDECDF-PAAC-IGGDCINTNGSYRCLCPQGH 853
Query 89 EAADAKTCK 97
+ C+
Sbjct 854 RLVGGRKCQ 862
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 31/64 (48%), Gaps = 2/64 (3%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
+ C NT GSY C C G+ G C+D+DEC+ C C N+ GS+ C C
Sbjct 834 IGGDCINTNGSYRCLCPQGHRLVG--GRKCQDIDECSQDPGLCLPHGACENLQGSYVCVC 891
Query 85 MAGF 88
GF
Sbjct 892 DEGF 895
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 41/90 (45%), Gaps = 5/90 (5%)
Query 6 RQCVFVPQGEASAAHSSTSLHASCANTEGSYVCTCNPGYEP-ASSDGHACKDVDECAAGT 64
R CV V + EA + C NT GSY C CN GY + G +C D++EC
Sbjct 609 RYCVDVNECEAEPCGPGKGI---CMNTGGSYNCHCNRGYRLHVGAGGRSCVDLNECTK-P 664
Query 65 AECHVSAQCVNVDGSFECTCMAGFEAADAK 94
C C+N G ++C C G+ ++
Sbjct 665 HLCGDGGFCINFPGHYKCNCYPGYRLKASR 694
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/70 (35%), Positives = 34/70 (48%), Gaps = 3/70 (4%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT-CMAG 87
C N G Y C C PGY +S C+D+DEC + C +C N GSF+C C G
Sbjct 673 CINFPGHYKCNCYPGYRLKASRPPICEDIDECRD-PSTC-PDGKCENKPGSFKCIACQPG 730
Query 88 FEAADAKTCK 97
+ + C+
Sbjct 731 YRSQGGGACR 740
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 30/64 (46%), Gaps = 3/64 (4%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHV--SAQCVNVDGSFECTC 84
C G VC C G++ +S C D+DEC + S +CVN GSF C C
Sbjct 1176 GRCVPRPGGAVCECPGGFQLDASRAR-CVDIDECRELNQRGLLCKSERCVNTSGSFRCVC 1234
Query 85 MAGF 88
AGF
Sbjct 1235 KAGF 1238
Score = 34.3 bits (77), Expect = 0.092, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 23/34 (67%), Gaps = 1/34 (2%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDE 59
H +C N +GSYVC C+ G+ + D H C++V++
Sbjct 877 HGACENLQGSYVCVCDEGFT-LTQDQHGCEEVEQ 909
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 28/67 (41%), Gaps = 8/67 (11%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAA---GTAECHVSAQCVNVDGSFEC 82
H C Y C CN GY + C DV+EC A G + C+N GS+ C
Sbjct 584 HGQCVPGPSDYSCHCNAGYR-SHPQHRYCVDVNECEAEPCGPGK----GICMNTGGSYNC 638
Query 83 TCMAGFE 89
C G+
Sbjct 639 HCNRGYR 645
Score = 32.3 bits (72), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 55 KDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
+D+DEC AE +CVN +EC C GF
Sbjct 989 RDIDECILFGAEICKEGKCVNTQPGYECYCKQGF 1022
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 31/70 (44%), Gaps = 7/70 (10%)
Query 27 ASCANTEGSYVCTCNPGYEPASSDGHA--CKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
C NT+ Y C C G+ DG+ C DVDEC + C + C N G + C C
Sbjct 1005 GKCVNTQPGYECYCKQGF---YYDGNLLECVDVDECLD-ESNCR-NGVCENTRGGYRCAC 1059
Query 85 MAGFEAADAK 94
E + A+
Sbjct 1060 TPPAEYSPAQ 1069
Score = 28.5 bits (62), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 27/56 (48%), Gaps = 4/56 (7%)
Query 40 CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGFEAADAKT 95
C GY+ +S C+D++ECA H C+N GS+ C C G ++T
Sbjct 338 CPQGYKRLNST--HCQDINECAMPGMCRH--GDCLNNPGSYRCVCPPGHSLGPSRT 389
> mmu:14114 Fbln1; fibulin 1
Length=705
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 30/71 (42%), Positives = 44/71 (61%), Gaps = 2/71 (2%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC-MAG 87
CAN GSY C C GY+ + DG C+D+DECA T S +C+N+ GSF+C+C +G
Sbjct 456 CANVYGSYQCYCRRGYQLSDVDGVTCEDIDECALPTGGHICSYRCINIPGSFQCSCPSSG 515
Query 88 FEAA-DAKTCK 97
+ A + + C+
Sbjct 516 YRLAPNGRNCQ 526
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 40/73 (54%), Gaps = 5/73 (6%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFEC----TC 84
C +T +C+C GY+ SDG +C+D++EC G+ C + C+N GSF C +C
Sbjct 192 CRDTGDEVICSCFVGYQ-LQSDGVSCEDINECITGSHNCRLGESCINTVGSFRCQRDSSC 250
Query 85 MAGFEAADAKTCK 97
G+E + CK
Sbjct 251 GTGYELTEDNNCK 263
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 34/66 (51%), Gaps = 6/66 (9%)
Query 28 SCANTEGSYVC-----TCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFEC 82
+C NTEGSY C C GY + +G C DVDEC+ C C+N GSF C
Sbjct 326 TCINTEGSYTCQKNVPNCGRGYH-LNEEGTRCVDVDECSPPAEPCGKGHHCLNSPGSFRC 384
Query 83 TCMAGF 88
C AGF
Sbjct 385 ECKAGF 390
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 39/66 (59%), Gaps = 5/66 (7%)
Query 28 SCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAG 87
C NT GS+ C+C+ G+ S DG +C+DV+EC S +C NV GS++C C G
Sbjct 416 KCENTPGSFHCSCSAGFR-LSVDGRSCEDVNECLNSPC----SQECANVYGSYQCYCRRG 470
Query 88 FEAADA 93
++ +D
Sbjct 471 YQLSDV 476
Score = 47.8 bits (112), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 23/56 (41%), Positives = 32/56 (57%), Gaps = 2/56 (3%)
Query 28 SCANTEGSYVCTC-NPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFEC 82
C N GS+ C+C + GY A +G C+D+DEC G C ++ C N+ GSF C
Sbjct 499 RCINIPGSFQCSCPSSGYRLAP-NGRNCQDIDECVTGIHNCSINETCFNIQGSFRC 553
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 36/73 (49%), Gaps = 10/73 (13%)
Query 24 SLHASCANTEGSYVC----TCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGS 79
L SC NT GS+ C +C GYE ++ + CKD+DEC G C C N GS
Sbjct 230 RLGESCINTVGSFRCQRDSSCGTGYE--LTEDNNCKDIDECETGIHNCPPDFICQNTLGS 287
Query 80 FECT----CMAGF 88
F C C +GF
Sbjct 288 FRCRPKLQCKSGF 300
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 36/72 (50%), Gaps = 7/72 (9%)
Query 29 CANTEGSYVCTCNPGYEPASSDG--HACKDVDECAAGTAECHVSAQCVNVDGSFECTCMA 86
C N+ GS+ C C G+ DG C D++EC +C N GSF C+C A
Sbjct 375 CLNSPGSFRCECKAGF---YFDGISRTCVDINECQRYPGRL-CGHKCENTPGSFHCSCSA 430
Query 87 GFE-AADAKTCK 97
GF + D ++C+
Sbjct 431 GFRLSVDGRSCE 442
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 30/58 (51%), Gaps = 6/58 (10%)
Query 29 CANTEGSYVCT----CNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFEC 82
C NT GS+ C C G+ + C D++EC + +A C V C+N +GS+ C
Sbjct 281 CQNTLGSFRCRPKLQCKSGFIQDALGN--CIDINECLSISAPCPVGQTCINTEGSYTC 336
> dre:792970 CD97 antigen-like; K08446 CD97 molecule
Length=670
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 36/64 (56%), Gaps = 2/64 (3%)
Query 25 LHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTC 84
LH SC NT GS+VC C+PG+ +G C D++EC+ C + C+N G F C C
Sbjct 39 LHRSCFNTPGSFVCKCSPGFREL--EGGMCADINECSESPDVCGTNGDCINNPGGFSCRC 96
Query 85 MAGF 88
GF
Sbjct 97 HQGF 100
Score = 35.8 bits (81), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 30/68 (44%), Gaps = 4/68 (5%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT GSYVC C G++ + +D ++ C + C N GSF C C GF
Sbjct 3 CENTFGSYVCVCPTGHQWKNEAECEDED----ECLSSPCGLHRSCFNTPGSFVCKCSPGF 58
Query 89 EAADAKTC 96
+ C
Sbjct 59 RELEGGMC 66
> dre:790930 nell2; zgc:158375
Length=811
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 28/65 (43%), Positives = 37/65 (56%), Gaps = 3/65 (4%)
Query 26 HASCANTEGSYVCTCNPGY---EPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFEC 82
HA+C N G Y C C GY E S++G +C+D+DEC G + C C N+DG F+C
Sbjct 567 HATCVNLPGWYHCECRDGYHDNEVFSANGESCRDIDECRTGRSTCANDTVCFNLDGGFDC 626
Query 83 TCMAG 87
C G
Sbjct 627 RCPHG 631
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 37/73 (50%), Gaps = 0/73 (0%)
Query 24 SLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECT 83
S ++ C N + C C G+ P D C+D+DECA G C + +CVN GSF C
Sbjct 406 SENSDCVNLDAGASCGCKNGFRPLRLDSAYCEDIDECAEGRHYCRENTECVNTAGSFMCV 465
Query 84 CMAGFEAADAKTC 96
C GF D +C
Sbjct 466 CHTGFIRIDDYSC 478
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 36/63 (57%), Gaps = 2/63 (3%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+ C NT GS++C C+ G+ D ++C + DECA+G +C +A C N G C+C
Sbjct 452 NTECVNTAGSFMCVCHTGF--IRIDDYSCTEHDECASGQHDCDENALCFNTVGGHSCSCK 509
Query 86 AGF 88
G+
Sbjct 510 PGY 512
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 29/109 (26%), Positives = 45/109 (41%), Gaps = 40/109 (36%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACK------------------------------ 55
+A C NT G + C+C PGY S +G C+
Sbjct 494 NALCFNTVGGHSCSCKPGY---SGNGTVCRALCDGRCLNGGSCASPNVCVCVQGFSGQNC 550
Query 56 --DVDECAAGTAECHVSAQCVNVDGSFECTCMAGFE-----AADAKTCK 97
D+DEC+ G +C A CVN+ G + C C G+ +A+ ++C+
Sbjct 551 ETDIDECSEGLVQCAAHATCVNLPGWYHCECRDGYHDNEVFSANGESCR 599
> mmu:170757 Eltd1, 1110033N21Rik, ETL1, Etl; EGF, latrophilin
seven transmembrane domain containing 1; K04595 latrophilin
and seven transmembrane domain containing 1
Length=739
Score = 57.8 bits (138), Expect = 9e-09, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 41/77 (53%), Gaps = 8/77 (10%)
Query 26 HASCANTEGSYVCTCNPGYEPAS-------SDGHACKDVDECAAGTAECHVSAQCVNVDG 78
HA C N G + C C GY+ A+ +DG C+D+DEC+ + C A C NV+G
Sbjct 71 HAVCENVNGGFSCFCREGYQTATGKSQFTPNDGSYCQDIDECSESSV-CGDHAVCENVNG 129
Query 79 SFECTCMAGFEAADAKT 95
F C C G++ A K+
Sbjct 130 GFSCFCREGYQTATGKS 146
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 24/70 (34%), Positives = 36/70 (51%), Gaps = 3/70 (4%)
Query 26 HASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCM 85
+A C G C C+ GY + + C+D+DEC+ + C A C NV+G F C C
Sbjct 30 NAKCEVHNGVEACFCSQGY--SGNGVTICEDIDECSESSV-CGDHAVCENVNGGFSCFCR 86
Query 86 AGFEAADAKT 95
G++ A K+
Sbjct 87 EGYQTATGKS 96
> dre:494044 efemp2, wu:fb22f10, wu:fc49d09, zgc:103659; EGF-containing
fibulin-like extracellular matrix protein 2
Length=440
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 28/69 (40%), Positives = 38/69 (55%), Gaps = 3/69 (4%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NT G+++C C GYE DG AC D+DEC+ + C QCVN G F C C G+
Sbjct 214 CYNTYGTFLCRCEQGYE-LGPDGFACNDIDECSYSSYLCQY--QCVNEHGKFSCVCPEGY 270
Query 89 EAADAKTCK 97
+ + C+
Sbjct 271 QLLGTRLCQ 279
Score = 48.5 bits (114), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 28/63 (44%), Positives = 33/63 (52%), Gaps = 7/63 (11%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C NTEGS+ C C GY G C D+DEC + +CVNV GSF C C GF
Sbjct 137 CFNTEGSFSCQCPDGYRKV---GTECIDIDECRYR----YCQHRCVNVPGSFSCQCEPGF 189
Query 89 EAA 91
+ A
Sbjct 190 QLA 192
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 34/74 (45%), Gaps = 3/74 (4%)
Query 18 AAHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVD 77
AA + S N +C PG+EP +C DVDEC + +C S QC N +
Sbjct 85 AAEPPNQIDPSLENRVSEPFNSCPPGFEPQQD---SCVDVDECERDSHDCQPSQQCFNTE 141
Query 78 GSFECTCMAGFEAA 91
GSF C C G+
Sbjct 142 GSFSCQCPDGYRKV 155
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/63 (42%), Positives = 35/63 (55%), Gaps = 4/63 (6%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
C N GS+ C C PG++ A ++ +C DVDEC G A C S C N G+F C C G+
Sbjct 174 CVNVPGSFSCQCEPGFQLAGNN-RSCIDVDECGMG-APC--SQWCYNTYGTFLCRCEQGY 229
Query 89 EAA 91
E
Sbjct 230 ELG 232
Score = 41.2 bits (95), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 36/79 (45%), Gaps = 10/79 (12%)
Query 19 AHSSTSLHASCANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDG 78
++SS C N G + C C GY+ + C+D++EC G +C CVN+ G
Sbjct 245 SYSSYLCQYQCVNEHGKFSCVCPEGYQLLGT--RLCQDINECETGAHQCSDGQTCVNIHG 302
Query 79 SFECTCMAGFEAADAKTCK 97
+C D K+C+
Sbjct 303 GHKC--------VDNKSCQ 313
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 27/63 (42%), Gaps = 3/63 (4%)
Query 22 STSLHASCAN--TEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGS 79
S LH + + TE C GY S H CKD++EC T C +C N G
Sbjct 14 SVFLHRAISQPPTETDTYTECTDGYHWGSQTQH-CKDINECETITEACKGEMKCFNHYGG 72
Query 80 FEC 82
+ C
Sbjct 73 YLC 75
> dre:449806 fbln5, zgc:103575; fibulin 5
Length=477
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 36/70 (51%), Positives = 43/70 (61%), Gaps = 6/70 (8%)
Query 29 CANTEGSYVCTCNPGYEPASSDGHACKDVDECAAGTAECHVSAQCVNVDGSFECTCMAGF 88
CAN GSY C+C+PG+ ++D C+DVDEC T C S C+N GSF CTC GF
Sbjct 211 CANVPGSYSCSCSPGFL-LNTDARTCQDVDECV--TDPC--SHGCLNTYGSFMCTCDEGF 265
Query 89 E-AADAKTCK 97
E AAD TC
Sbjct 266 ELAADDTTCN 275
Lambda K H
0.318 0.127 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2055684140
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40