bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5783_orf2
Length=113
Score E
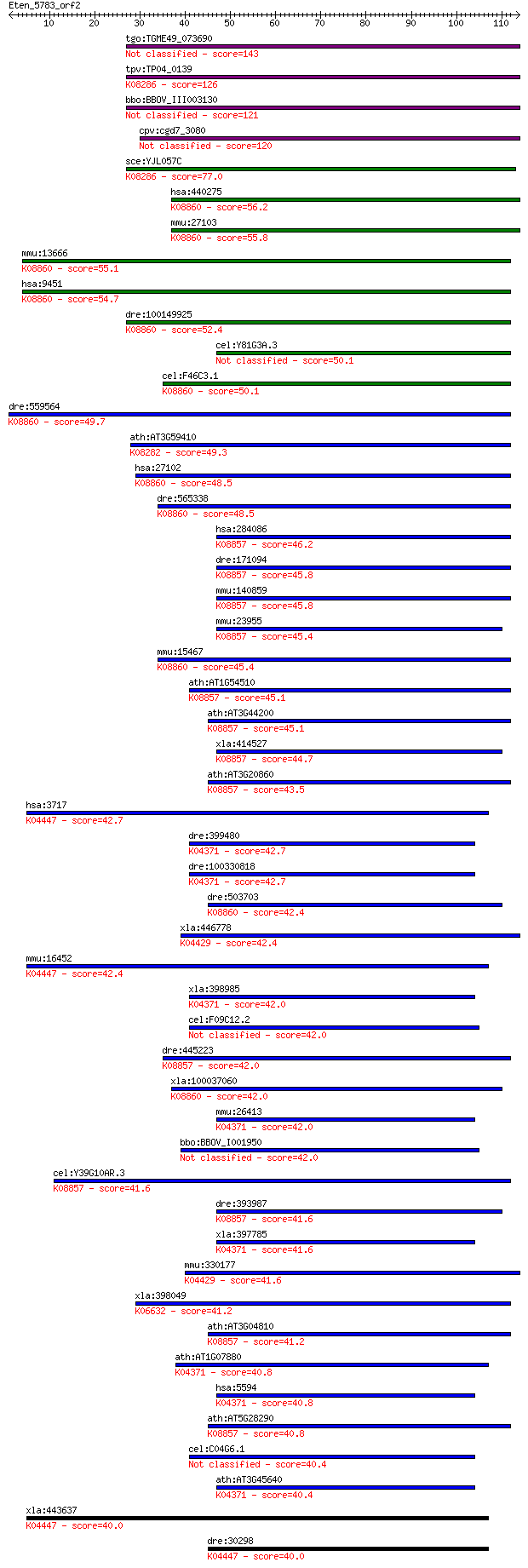
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_073690 serine/threonine protein kinase, putative (E... 143 1e-34
tpv:TP04_0139 serine/threonine protein kinase (EC:2.7.1.37); K... 126 1e-29
bbo:BBOV_III003130 17.m07300; protein kinase domain containing... 121 5e-28
cpv:cgd7_3080 hypothetical protein 120 2e-27
sce:YJL057C IKS1; Iks1p (EC:2.7.11.1); K08286 protein-serine/t... 77.0 1e-14
hsa:440275 EIF2AK4, GCN2, KIAA1338; eukaryotic translation ini... 56.2 2e-08
mmu:27103 Eif2ak4, 2610011M03, GCN2, MGCN2; eukaryotic transla... 55.8 3e-08
mmu:13666 Eif2ak3, AI427929, Pek, Perk; eukaryotic translation... 55.1 5e-08
hsa:9451 EIF2AK3, DKFZp781H1925, PEK, PERK, WRS; eukaryotic tr... 54.7 7e-08
dre:100149925 eif2ak4, GCN2; eukaryotic translation initiation... 52.4 4e-07
cel:Y81G3A.3 hypothetical protein 50.1 2e-06
cel:F46C3.1 pek-1; human PERK kinase homolog family member (pe... 50.1 2e-06
dre:559564 eif2ak3, MGC152949, PEK, zgc:152949; eukaryotic tra... 49.7 2e-06
ath:AT3G59410 protein kinase family protein; K08282 non-specif... 49.3 3e-06
hsa:27102 EIF2AK1, HCR, HRI, KIAA1369; eukaryotic translation ... 48.5 5e-06
dre:565338 eif2ak1, MGC153232, wu:fj97h09, zgc:153232; eukaryo... 48.5 5e-06
hsa:284086 NEK8, JCK, MGC138445, NEK12A, NPHP9; NIMA (never in... 46.2 2e-05
dre:171094 nek8; NIMA (never in mitosis gene a)-related kinase... 45.8 3e-05
mmu:140859 Nek8, 4632401F23Rik, jck; NIMA (never in mitosis ge... 45.8 4e-05
mmu:23955 Nek4; NIMA (never in mitosis gene a)-related express... 45.4 4e-05
mmu:15467 Eif2ak1, Hri, MGC118066; eukaryotic translation init... 45.4 5e-05
ath:AT1G54510 ATNEK1; ATNEK1 (ARABIDOPSIS THALIANA NIMA-RELATE... 45.1 6e-05
ath:AT3G44200 NEK6; NEK6 ('NIMA (NEVER IN MITOSIS, GENE A)-REL... 45.1 6e-05
xla:414527 nek4, MGC81305; NIMA (never in mitosis gene a)-rela... 44.7 7e-05
ath:AT3G20860 ATNEK5; ATNEK5 (NIMA-RELATED KINASE5); ATP bindi... 43.5 2e-04
hsa:3717 JAK2, JTK10; Janus kinase 2 (EC:2.7.10.2); K04447 Jan... 42.7 3e-04
dre:399480 mapk3, ERK1, fi06b09, wu:fi06b09, zERK1; mitogen-ac... 42.7 3e-04
dre:100330818 mitogen-activated protein kinase 3-like; K04371 ... 42.7 3e-04
dre:503703 pkz, MGC136657, MGC158143, zgc:136657; protein kina... 42.4 3e-04
xla:446778 taok3, MGC80412; TAO kinase 3 (EC:2.7.11.1); K04429... 42.4 3e-04
mmu:16452 Jak2, AI504024, C81284, Fd17; Janus kinase 2 (EC:2.7... 42.4 4e-04
xla:398985 mapk1-a, MGC68934, erk, erk2, mapk, mapk1, mapk1a, ... 42.0 4e-04
cel:F09C12.2 hypothetical protein 42.0 5e-04
dre:445223 nek7, MGC92175, wu:fj68f02, zgc:100962, zgc:92175; ... 42.0 5e-04
xla:100037060 eif2ak2, eif2ak1, pkr, pkr1, prkr; eukaryotic tr... 42.0 5e-04
mmu:26413 Mapk1, 9030612K14Rik, AA407128, AU018647, C78273, ER... 42.0 5e-04
bbo:BBOV_I001950 19.m02272; protein kinase domain containing p... 42.0 5e-04
cel:Y39G10AR.3 nekl-1; NEK (NEver in mitosis Kinase) Like fami... 41.6 6e-04
dre:393987 nek4, MGC64176, zgc:64176; NIMA (never in mitosis g... 41.6 7e-04
xla:397785 mapk1-b, Xp42, erk, erk2, mapk, mapk1b, mpk1; mitog... 41.6 7e-04
mmu:330177 Taok3, 2900006A08Rik, A130052D22, A430105I05Rik; TA... 41.6 7e-04
xla:398049 wee1-a, MGC81696, wee1a, xe-wee1a, xwee1; wee1 homo... 41.2 9e-04
ath:AT3G04810 ATNEK2; ATNEK2 (NIMA-RELATED KINASE 2); ATP bind... 41.2 0.001
ath:AT1G07880 ATMPK13; MAP kinase/ kinase; K04371 extracellula... 40.8 0.001
hsa:5594 MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2,... 40.8 0.001
ath:AT5G28290 ATNEK3; ATNEK3 (NIMA-RELATED KINASE3); ATP bindi... 40.8 0.001
cel:C04G6.1 mpk-2; MAP Kinase family member (mpk-2) 40.4 0.001
ath:AT3G45640 ATMPK3; ATMPK3 (ARABIDOPSIS THALIANA MITOGEN-ACT... 40.4 0.002
xla:443637 jak2, MGC114825; Janus kinase 2 (a protein tyrosine... 40.0 0.002
dre:30298 jak2b, JAK2bCA, jak2; Janus kinase 2b (EC:2.7.10.2 2... 40.0 0.002
> tgo:TGME49_073690 serine/threonine protein kinase, putative
(EC:2.7.12.2)
Length=482
Score = 143 bits (360), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 62/87 (71%), Positives = 76/87 (87%), Gaps = 0/87 (0%)
Query 27 IPREFLVTGYYSRFFLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQR 86
IP E L+TGYY+RFF+E+Q++G G++G + LC H LDEL LGE+AVKKLPVGDD+HWL +
Sbjct 63 IPAELLITGYYNRFFIEKQKLGSGSYGQVYLCTHILDELTLGEYAVKKLPVGDDKHWLAK 122
Query 87 ALREVKIRERLSHPNIVDYKHSWLEMH 113
L+EVKIRE+L HPNIVDYKHSWLEMH
Sbjct 123 MLQEVKIREKLHHPNIVDYKHSWLEMH 149
> tpv:TP04_0139 serine/threonine protein kinase (EC:2.7.1.37);
K08286 protein-serine/threonine kinase [EC:2.7.11.-]
Length=511
Score = 126 bits (317), Expect = 1e-29, Method: Composition-based stats.
Identities = 50/87 (57%), Positives = 70/87 (80%), Gaps = 0/87 (0%)
Query 27 IPREFLVTGYYSRFFLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQR 86
IP+ L+TGYY RFF+E ++G G++G + C H LDEL+LGE+AVKK+P+GDD WL++
Sbjct 164 IPKYLLITGYYKRFFIEGPKLGSGSYGHVYNCIHILDELVLGEYAVKKIPIGDDMEWLKK 223
Query 87 ALREVKIRERLSHPNIVDYKHSWLEMH 113
A++E+KIRE + H N+VDY HSWLEM+
Sbjct 224 AIKEIKIRENIKHKNVVDYNHSWLEMY 250
> bbo:BBOV_III003130 17.m07300; protein kinase domain containing
protein
Length=380
Score = 121 bits (304), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 53/87 (60%), Positives = 69/87 (79%), Gaps = 0/87 (0%)
Query 27 IPREFLVTGYYSRFFLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQR 86
IP + LVTGYY RFF+E+ ++G G++G + C H +D L LGE+A+KKLPVGDDR WL++
Sbjct 42 IPPDLLVTGYYHRFFIEKMKLGSGSYGHVYHCVHVIDGLSLGEYAIKKLPVGDDRQWLRK 101
Query 87 ALREVKIRERLSHPNIVDYKHSWLEMH 113
+REVK+RE L H NIVDY HSWLEM+
Sbjct 102 MIREVKVREMLRHQNIVDYNHSWLEMY 128
> cpv:cgd7_3080 hypothetical protein
Length=665
Score = 120 bits (300), Expect = 2e-27, Method: Composition-based stats.
Identities = 49/84 (58%), Positives = 67/84 (79%), Gaps = 0/84 (0%)
Query 30 EFLVTGYYSRFFLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRALR 89
+ L+TGYY +FF+E +++G G+FG + LC H LD + L ++A+KK+PVGDD++WL LR
Sbjct 254 DLLMTGYYEKFFVEIRKLGSGSFGQVYLCAHVLDGITLAKYAIKKVPVGDDKNWLSAVLR 313
Query 90 EVKIRERLSHPNIVDYKHSWLEMH 113
EVKIRE L HPNIV Y+HSWLEM+
Sbjct 314 EVKIRELLHHPNIVQYRHSWLEMY 337
> sce:YJL057C IKS1; Iks1p (EC:2.7.11.1); K08286 protein-serine/threonine
kinase [EC:2.7.11.-]
Length=667
Score = 77.0 bits (188), Expect = 1e-14, Method: Composition-based stats.
Identities = 34/88 (38%), Positives = 51/88 (57%), Gaps = 2/88 (2%)
Query 27 IPREFLVTGYYSRFFLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQR 86
IP + + GY+ +FF +G GA G++ H + LG FA+KK+P+G+D W +
Sbjct 159 IPDDLFIPGYFRKFFKILSLLGNGARGSVYKVVHTIGNTELGVFALKKIPIGNDMEWFNK 218
Query 87 ALREVKIRERLSH--PNIVDYKHSWLEM 112
+REVK L+H N++ Y H WLEM
Sbjct 219 CIREVKALSSLTHKSANLITYNHVWLEM 246
> hsa:440275 EIF2AK4, GCN2, KIAA1338; eukaryotic translation initiation
factor 2 alpha kinase 4 (EC:2.7.11.1); K08860 eukaryotic
translation initiation factor 2-alpha kinase [EC:2.7.11.1]
Length=1649
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 47/80 (58%), Gaps = 6/80 (7%)
Query 37 YSRFFLE---RQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRALREVKI 93
+SR+F+E Q +G+GAFG + ++KLD +AVK++P+ +R EV +
Sbjct 583 FSRYFIEFEELQLLGKGAFGAVIKVQNKLDGCC---YAVKRIPINPASRQFRRIKGEVTL 639
Query 94 RERLSHPNIVDYKHSWLEMH 113
RL H NIV Y ++W+E H
Sbjct 640 LSRLHHENIVRYYNAWIERH 659
> mmu:27103 Eif2ak4, 2610011M03, GCN2, MGCN2; eukaryotic translation
initiation factor 2 alpha kinase 4 (EC:2.7.11.1); K08860
eukaryotic translation initiation factor 2-alpha kinase
[EC:2.7.11.1]
Length=1536
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 47/80 (58%), Gaps = 6/80 (7%)
Query 37 YSRFFLE---RQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRALREVKI 93
+SR+F+E Q +G+GAFG + ++KLD +AVK++P+ +R EV +
Sbjct 470 FSRYFIEFEELQLLGKGAFGAVIKVQNKLDGCC---YAVKRIPINPASRHFRRIKGEVTL 526
Query 94 RERLSHPNIVDYKHSWLEMH 113
RL H NIV Y ++W+E H
Sbjct 527 LSRLHHENIVRYYNAWIERH 546
> mmu:13666 Eif2ak3, AI427929, Pek, Perk; eukaryotic translation
initiation factor 2 alpha kinase 3 (EC:2.7.11.1); K08860
eukaryotic translation initiation factor 2-alpha kinase [EC:2.7.11.1]
Length=1114
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 37/122 (30%), Positives = 64/122 (52%), Gaps = 18/122 (14%)
Query 4 LFNPYLRHRQRR---LQVEETADYEPIP--------REFLVTGYYSRF---FLERQQIGR 49
LF+P HRQR+ Q + + Y+ + + +GY SR+ F Q +GR
Sbjct 539 LFHPQ-PHRQRKESETQCQTESKYDSVSADVSDNSWNDMKYSGYVSRYLTDFEPIQCMGR 597
Query 50 GAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRALREVKIRERLSHPNIVDYKHSW 109
G FG + ++K+D+ +A+K++ + + ++ +REVK +L HP IV Y ++W
Sbjct 598 GGFGVVFEAKNKVDDC---NYAIKRIRLPNRELAREKVMREVKALAKLEHPGIVRYFNAW 654
Query 110 LE 111
LE
Sbjct 655 LE 656
> hsa:9451 EIF2AK3, DKFZp781H1925, PEK, PERK, WRS; eukaryotic
translation initiation factor 2-alpha kinase 3 (EC:2.7.11.1);
K08860 eukaryotic translation initiation factor 2-alpha kinase
[EC:2.7.11.1]
Length=1116
Score = 54.7 bits (130), Expect = 7e-08, Method: Composition-based stats.
Identities = 38/122 (31%), Positives = 64/122 (52%), Gaps = 18/122 (14%)
Query 4 LFNPYLRHRQRR---LQVEETADYEPIPRE--------FLVTGYYSRF---FLERQQIGR 49
LF+P+ HRQR+ Q + Y+ + E +GY SR+ F Q +GR
Sbjct 543 LFHPH-PHRQRKESETQCQTENKYDSVSGEANDSSWNDIKNSGYISRYLTDFEPIQCLGR 601
Query 50 GAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRALREVKIRERLSHPNIVDYKHSW 109
G FG + ++K+D+ +A+K++ + + ++ +REVK +L HP IV Y ++W
Sbjct 602 GGFGVVFEAKNKVDDC---NYAIKRIRLPNRELAREKVMREVKALAKLEHPGIVRYFNAW 658
Query 110 LE 111
LE
Sbjct 659 LE 660
> dre:100149925 eif2ak4, GCN2; eukaryotic translation initiation
factor 2 alpha kinase 4; K08860 eukaryotic translation initiation
factor 2-alpha kinase [EC:2.7.11.1]
Length=1483
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 51/96 (53%), Gaps = 14/96 (14%)
Query 27 IPREFLVTG--------YYSRF---FLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKL 75
+PR L++ +SR+ F E Q +G+GAFG + ++KLD +AVK++
Sbjct 500 VPRSLLLSAPFSTEVQKQFSRYYNEFSELQLLGKGAFGAVIKVQNKLDGCY---YAVKRI 556
Query 76 PVGDDRHWLQRALREVKIRERLSHPNIVDYKHSWLE 111
V +R EV + RL+H NIV Y ++W+E
Sbjct 557 QVNPASKQFRRIKGEVTLLSRLNHENIVRYYNAWIE 592
> cel:Y81G3A.3 hypothetical protein
Length=1696
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 23/66 (34%), Positives = 40/66 (60%), Gaps = 4/66 (6%)
Query 47 IGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRAL-REVKIRERLSHPNIVDY 105
+GRG FG + L +K+D ++A+K++P+ L R + +E K +L+HPN+V Y
Sbjct 497 LGRGGFGDVVLVRNKMDST---DYAIKRIPLNAKSDKLNRKIAKEAKFFAKLNHPNMVRY 553
Query 106 KHSWLE 111
++W E
Sbjct 554 YYAWAE 559
> cel:F46C3.1 pek-1; human PERK kinase homolog family member (pek-1);
K08860 eukaryotic translation initiation factor 2-alpha
kinase [EC:2.7.11.1]
Length=1077
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 27/80 (33%), Positives = 41/80 (51%), Gaps = 6/80 (7%)
Query 35 GYYSRF---FLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRALREV 91
G+ SRF F ++ IG G FG + + D + E+AVK++ V D+ R LRE
Sbjct 595 GFTSRFANEFEVKKVIGHGGFGVVFRAQSITD---MNEYAVKRIAVADNDKARNRVLREA 651
Query 92 KIRERLSHPNIVDYKHSWLE 111
+ HP I+ Y ++W E
Sbjct 652 RALAMFDHPGIIRYFYAWEE 671
> dre:559564 eif2ak3, MGC152949, PEK, zgc:152949; eukaryotic translation
initiation factor 2-alpha kinase 3 (EC:2.7.11.1);
K08860 eukaryotic translation initiation factor 2-alpha kinase
[EC:2.7.11.1]
Length=1099
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 32/120 (26%), Positives = 61/120 (50%), Gaps = 12/120 (10%)
Query 1 PPALFNPYLRHRQRRLQVEETADYEPI-PREFLVTG-----YYSRF---FLERQQIGRGA 51
PP ++ + + + Q + + + P+E +VT Y SR+ F Q +GRG
Sbjct 563 PPTTYSRQRKESETQCQTDTKFEADVTEPKEVVVTDMRPCEYVSRYLTDFEPVQCLGRGG 622
Query 52 FGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRALREVKIRERLSHPNIVDYKHSWLE 111
FG + +K+D+ +A+K++ + + ++ +REVK +L HP I+ Y ++W E
Sbjct 623 FGVVFEARNKVDDC---NYAIKRIRLPNRELAREKVMREVKALAKLEHPGIIRYFNAWQE 679
> ath:AT3G59410 protein kinase family protein; K08282 non-specific
serine/threonine protein kinase [EC:2.7.11.1]
Length=1241
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 30/85 (35%), Positives = 46/85 (54%), Gaps = 4/85 (4%)
Query 28 PREFLVTGYYSRFFLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQ-R 86
P L + Y F E + +G+G FG + LC++KLD ++AVKK+ + D + R
Sbjct 412 PNASLPSSRYLNDFEELKPLGQGGFGHVVLCKNKLDG---RQYAVKKIRLKDKEIPVNSR 468
Query 87 ALREVKIRERLSHPNIVDYKHSWLE 111
+REV RL H ++V Y +W E
Sbjct 469 IVREVATLSRLQHQHVVRYYQAWFE 493
> hsa:27102 EIF2AK1, HCR, HRI, KIAA1369; eukaryotic translation
initiation factor 2-alpha kinase 1 (EC:2.7.11.1); K08860 eukaryotic
translation initiation factor 2-alpha kinase [EC:2.7.11.1]
Length=629
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 30/88 (34%), Positives = 49/88 (55%), Gaps = 9/88 (10%)
Query 29 REFLVTGYYSRFFLERQQI---GRGAFGTICLCEHKLDELLLGEF-AVKKLPV-GDDRHW 83
RE + SR+ E +++ G+G +G + +KLD G++ A+KK+ + G +
Sbjct 152 REVALEAQTSRYLNEFEELAILGKGGYGRVYKVRNKLD----GQYYAIKKILIKGATKTV 207
Query 84 LQRALREVKIRERLSHPNIVDYKHSWLE 111
+ LREVK+ L HPNIV Y +W+E
Sbjct 208 CMKVLREVKVLAGLQHPNIVGYHTAWIE 235
> dre:565338 eif2ak1, MGC153232, wu:fj97h09, zgc:153232; eukaryotic
translation initiation factor 2-alpha kinase 1 (EC:2.7.11.1);
K08860 eukaryotic translation initiation factor 2-alpha
kinase [EC:2.7.11.1]
Length=621
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 29/79 (36%), Positives = 43/79 (54%), Gaps = 4/79 (5%)
Query 34 TGYYSRFFLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGD-DRHWLQRALREVK 92
T Y F E +G+G++G + +KLD E+AVKK+ + R + LREVK
Sbjct 139 TSRYLSEFEEIATLGKGSYGKVFRVTNKLDG---QEYAVKKILIKKVTRDDCMKVLREVK 195
Query 93 IRERLSHPNIVDYKHSWLE 111
+ L HP+IV Y +W+E
Sbjct 196 VLSSLQHPDIVGYHTAWME 214
> hsa:284086 NEK8, JCK, MGC138445, NEK12A, NPHP9; NIMA (never
in mitosis gene a)- related kinase 8 (EC:2.7.11.1); K08857 NIMA
(never in mitosis gene a)-related kinase [EC:2.7.11.1]
Length=692
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 24/66 (36%), Positives = 41/66 (62%), Gaps = 4/66 (6%)
Query 47 IGRGAFGTICLCEHKLDELLLGEFAVKKLPVGD-DRHWLQRALREVKIRERLSHPNIVDY 105
+GRGAFG + LC K D+ L+ +K++PV + Q A E ++ + L+HPN+++Y
Sbjct 10 VGRGAFGIVHLCLRKADQKLV---IIKQIPVEQMTKEERQAAQNECQVLKLLNHPNVIEY 66
Query 106 KHSWLE 111
++LE
Sbjct 67 YENFLE 72
> dre:171094 nek8; NIMA (never in mitosis gene a)-related kinase
8 (EC:2.7.11.1); K08857 NIMA (never in mitosis gene a)-related
kinase [EC:2.7.11.1]
Length=697
Score = 45.8 bits (107), Expect = 3e-05, Method: Composition-based stats.
Identities = 25/66 (37%), Positives = 39/66 (59%), Gaps = 4/66 (6%)
Query 47 IGRGAFGTICLCEHKLDELLLGEFAVKKLPVGD-DRHWLQRALREVKIRERLSHPNIVDY 105
+GRGAFG + LC + D L+ +K++PV R A E ++ + LSHPNI++Y
Sbjct 10 VGRGAFGIVHLCRRRTDSALV---ILKEIPVEQMTRDERLAAQNECQVLKLLSHPNIIEY 66
Query 106 KHSWLE 111
++LE
Sbjct 67 YENFLE 72
> mmu:140859 Nek8, 4632401F23Rik, jck; NIMA (never in mitosis
gene a)-related expressed kinase 8 (EC:2.7.11.1); K08857 NIMA
(never in mitosis gene a)-related kinase [EC:2.7.11.1]
Length=698
Score = 45.8 bits (107), Expect = 4e-05, Method: Composition-based stats.
Identities = 24/66 (36%), Positives = 41/66 (62%), Gaps = 4/66 (6%)
Query 47 IGRGAFGTICLCEHKLDELLLGEFAVKKLPVGD-DRHWLQRALREVKIRERLSHPNIVDY 105
+GRGAFG + LC K D+ L+ +K++PV + Q A E ++ + L+HPN+++Y
Sbjct 10 VGRGAFGIVHLCLRKADQKLV---ILKQIPVEQMTKEERQAAQNECQVLKLLNHPNVIEY 66
Query 106 KHSWLE 111
++LE
Sbjct 67 YENFLE 72
> mmu:23955 Nek4; NIMA (never in mitosis gene a)-related expressed
kinase 4 (EC:2.7.11.1); K08857 NIMA (never in mitosis gene
a)-related kinase [EC:2.7.11.1]
Length=792
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 39/64 (60%), Gaps = 4/64 (6%)
Query 47 IGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRAL-REVKIRERLSHPNIVDY 105
+GRG++G + L +H+ D ++ +KKL + + +RA +E ++ +L HPNIV Y
Sbjct 12 VGRGSYGEVTLVKHRRDG---KQYVIKKLNLRNASSRERRAAEQEAQLLSQLKHPNIVTY 68
Query 106 KHSW 109
K SW
Sbjct 69 KESW 72
> mmu:15467 Eif2ak1, Hri, MGC118066; eukaryotic translation initiation
factor 2 alpha kinase 1 (EC:2.7.11.1); K08860 eukaryotic
translation initiation factor 2-alpha kinase [EC:2.7.11.1]
Length=619
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 43/80 (53%), Gaps = 6/80 (7%)
Query 34 TGYYSRFFLERQQIGRGAFGTICLCEHKLDELLLGE-FAVKKLPV-GDDRHWLQRALREV 91
T Y F E +G+G +G + +KLD G+ +A+KK+ + + + LREV
Sbjct 160 TSRYLNEFEELAILGKGGYGRVYKVRNKLD----GQHYAIKKILIKSATKTDCMKVLREV 215
Query 92 KIRERLSHPNIVDYKHSWLE 111
K+ L HPNIV Y +W+E
Sbjct 216 KVLAGLQHPNIVGYHTAWIE 235
> ath:AT1G54510 ATNEK1; ATNEK1 (ARABIDOPSIS THALIANA NIMA-RELATED
SERINE/THREONINE KINASE 1); ATP binding / kinase/ protein
kinase/ protein serine/threonine kinase; K08857 NIMA (never
in mitosis gene a)-related kinase [EC:2.7.11.1]
Length=612
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 26/72 (36%), Positives = 45/72 (62%), Gaps = 6/72 (8%)
Query 41 FLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQR-ALREVKIRERLSH 99
FLE QIG+G+FG+ L HK ++ ++ +KK+ + +R A +E+++ ++ H
Sbjct 6 FLE--QIGKGSFGSALLVRHKHEK---KKYVLKKIRLARQTQRTRRSAHQEMELISKMRH 60
Query 100 PNIVDYKHSWLE 111
P IV+YK SW+E
Sbjct 61 PFIVEYKDSWVE 72
> ath:AT3G44200 NEK6; NEK6 ('NIMA (NEVER IN MITOSIS, GENE A)-RELATED
6'); ATP binding / kinase/ protein kinase/ protein serine/threonine
kinase; K08857 NIMA (never in mitosis gene a)-related
kinase [EC:2.7.11.1]
Length=956
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 24/68 (35%), Positives = 40/68 (58%), Gaps = 4/68 (5%)
Query 45 QQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQR-ALREVKIRERLSHPNIV 103
+QIGRGAFG L HK + ++ +KK+ + +R A +E+ + R+ HP IV
Sbjct 12 EQIGRGAFGAAILVHHKAER---KKYVLKKIRLARQTERCRRSAHQEMSLIARVQHPYIV 68
Query 104 DYKHSWLE 111
++K +W+E
Sbjct 69 EFKEAWVE 76
> xla:414527 nek4, MGC81305; NIMA (never in mitosis gene a)-related
kinase 4; K08857 NIMA (never in mitosis gene a)-related
kinase [EC:2.7.11.1]
Length=790
Score = 44.7 bits (104), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 38/64 (59%), Gaps = 4/64 (6%)
Query 47 IGRGAFGTICLCEHKLDELLLGEFAVKKLPVGD-DRHWLQRALREVKIRERLSHPNIVDY 105
+G+G++G + L H+ L +F +KKL + + R + A +E ++ RL HPNIV Y
Sbjct 16 VGKGSYGEVSLVRHRT---LGKQFVIKKLNLQNASRRERKAAEQEAQLLSRLKHPNIVAY 72
Query 106 KHSW 109
+ SW
Sbjct 73 RESW 76
> ath:AT3G20860 ATNEK5; ATNEK5 (NIMA-RELATED KINASE5); ATP binding
/ kinase/ protein kinase/ protein serine/threonine kinase/
protein tyrosine kinase; K08857 NIMA (never in mitosis gene
a)-related kinase [EC:2.7.11.1]
Length=427
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 25/68 (36%), Positives = 40/68 (58%), Gaps = 4/68 (5%)
Query 45 QQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQ-RALREVKIRERLSHPNIV 103
+QIGRGAFG+ L HK + ++ VKK+ + + A++E+ + +L P IV
Sbjct 19 EQIGRGAFGSAFLVIHKSERR---KYVVKKIRLAKQTERCKLAAIQEMSLISKLKSPYIV 75
Query 104 DYKHSWLE 111
+YK SW+E
Sbjct 76 EYKDSWVE 83
> hsa:3717 JAK2, JTK10; Janus kinase 2 (EC:2.7.10.2); K04447 Janus
kinase 2 [EC:2.7.10.2]
Length=1132
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 40/122 (32%), Positives = 56/122 (45%), Gaps = 21/122 (17%)
Query 5 FNPYLRHRQRRLQVEETADYEPIPREFLVT-------GYYSRF-------FLER-----Q 45
F P R R L T DYE + ++ G+ F F ER Q
Sbjct 794 FRPSFRAIIRDLNSLFTPDYELLTENDMLPNMRIGALGFSGAFEDRDPTQFEERHLKFLQ 853
Query 46 QIGRGAFGTICLCEHKLDELLLGEF-AVKKLPVGDDRHWLQRALREVKIRERLSHPNIVD 104
Q+G+G FG++ +C + + GE AVKKL + H L+ RE++I + L H NIV
Sbjct 854 QLGKGNFGSVEMCRYDPLQDNTGEVVAVKKLQHSTEEH-LRDFEREIEILKSLQHDNIVK 912
Query 105 YK 106
YK
Sbjct 913 YK 914
> dre:399480 mapk3, ERK1, fi06b09, wu:fi06b09, zERK1; mitogen-activated
protein kinase 3 (EC:2.7.11.1); K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=392
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 36/63 (57%), Gaps = 3/63 (4%)
Query 41 FLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRALREVKIRERLSHP 100
+ + Q IG GA+G +C ++++ A+KK+ + + + QR LRE+KI R H
Sbjct 56 YTDLQYIGEGAYGMVCSAFDNVNKI---RVAIKKISPFEHQTYCQRTLREIKILLRFHHE 112
Query 101 NIV 103
NI+
Sbjct 113 NII 115
> dre:100330818 mitogen-activated protein kinase 3-like; K04371
extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=392
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 36/63 (57%), Gaps = 3/63 (4%)
Query 41 FLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRALREVKIRERLSHP 100
+ + Q IG GA+G +C ++++ A+KK+ + + + QR LRE+KI R H
Sbjct 56 YTDLQYIGEGAYGMVCSAFDNVNKI---RVAIKKISPFEHQTYCQRTLREIKILLRFHHE 112
Query 101 NIV 103
NI+
Sbjct 113 NII 115
> dre:503703 pkz, MGC136657, MGC158143, zgc:136657; protein kinase
containing Z-DNA binding domains; K08860 eukaryotic translation
initiation factor 2-alpha kinase [EC:2.7.11.1]
Length=511
Score = 42.4 bits (98), Expect = 3e-04, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 36/65 (55%), Gaps = 9/65 (13%)
Query 45 QQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRALREVKIRERLSHPNIVD 104
+++G G +G +C +HK+D+ + +AVK++ + A EVK RL HPNIV
Sbjct 174 EELGDGGYGFVCKVKHKIDDKI---YAVKRVEFNSE------AEPEVKALARLDHPNIVR 224
Query 105 YKHSW 109
Y W
Sbjct 225 YFTCW 229
> xla:446778 taok3, MGC80412; TAO kinase 3 (EC:2.7.11.1); K04429
thousand and one amino acid protein kinase [EC:2.7.11.1]
Length=896
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 42/78 (53%), Gaps = 7/78 (8%)
Query 39 RFFLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGD---DRHWLQRALREVKIRE 95
R F++ +IG G+FG + + ++ AVKK+ + W Q ++EVK +
Sbjct 22 RIFVDLHEIGHGSFGAVYFATNSTTNEIV---AVKKMSYSGKQMNEKW-QDIIKEVKFLQ 77
Query 96 RLSHPNIVDYKHSWLEMH 113
+L HPN ++YK +L+ H
Sbjct 78 QLKHPNTIEYKGCYLKDH 95
> mmu:16452 Jak2, AI504024, C81284, Fd17; Janus kinase 2 (EC:2.7.10.2);
K04447 Janus kinase 2 [EC:2.7.10.2]
Length=1132
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 40/122 (32%), Positives = 56/122 (45%), Gaps = 21/122 (17%)
Query 5 FNPYLRHRQRRLQVEETADYEPIPREFLV-------TGYYSRF-------FLER-----Q 45
F P R R L T DYE + ++ G+ F F ER Q
Sbjct 794 FRPAFRAVIRDLNSLFTPDYELLTENDMLPNMRIGALGFSGAFEDRDPTQFEERHLKFLQ 853
Query 46 QIGRGAFGTICLCEHKLDELLLGEF-AVKKLPVGDDRHWLQRALREVKIRERLSHPNIVD 104
Q+G+G FG++ +C + + GE AVKKL + H L+ RE++I + L H NIV
Sbjct 854 QLGKGNFGSVEMCRYDPLQDNTGEVVAVKKLQHSTEEH-LRDFEREIEILKSLQHDNIVK 912
Query 105 YK 106
YK
Sbjct 913 YK 914
> xla:398985 mapk1-a, MGC68934, erk, erk2, mapk, mapk1, mapk1a,
mpk1, xp42; mitogen-activated protein kinase 1 (EC:2.7.11.24);
K04371 extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=361
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 35/63 (55%), Gaps = 3/63 (4%)
Query 41 FLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRALREVKIRERLSHP 100
++ IG GA+G +C ++++ A+KK+ + + + QR LRE+KI R H
Sbjct 28 YINLAYIGEGAYGMVCSAHDNVNKV---RVAIKKISPFEHQTYCQRTLREIKILLRFKHE 84
Query 101 NIV 103
NI+
Sbjct 85 NII 87
> cel:F09C12.2 hypothetical protein
Length=418
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 39/65 (60%), Gaps = 4/65 (6%)
Query 41 FLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLP-VGDDRHWLQRALREVKIRERLSH 99
+L ++ +G GAFG +C +D L + A+KK+ V ++ + ALRE++I LSH
Sbjct 44 YLAQENLGAGAFGVVC---RAIDSKLNKQVAIKKITRVFKNQSTAKCALREIRITRELSH 100
Query 100 PNIVD 104
NI++
Sbjct 101 ENIIN 105
> dre:445223 nek7, MGC92175, wu:fj68f02, zgc:100962, zgc:92175;
NIMA (never in mitosis gene a)-related kinase 7 (EC:2.7.11.1);
K08857 NIMA (never in mitosis gene a)-related kinase [EC:2.7.11.1]
Length=297
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 47/81 (58%), Gaps = 8/81 (9%)
Query 35 GYYS--RFFLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGD--DRHWLQRALRE 90
GY S F +E++ IGRG F + + LD ++ A+KK+ + D D Q ++E
Sbjct 23 GYNSLANFAIEKK-IGRGQFSEVYRATYVLDHTIV---ALKKIQIFDLMDAKARQDCIKE 78
Query 91 VKIRERLSHPNIVDYKHSWLE 111
+ + ++L+HPN++ Y S++E
Sbjct 79 IDLLKQLNHPNVIKYHASFIE 99
> xla:100037060 eif2ak2, eif2ak1, pkr, pkr1, prkr; eukaryotic
translation initiation factor 2-alpha kinase 2; K08860 eukaryotic
translation initiation factor 2-alpha kinase [EC:2.7.11.1]
Length=578
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 23/73 (31%), Positives = 39/73 (53%), Gaps = 8/73 (10%)
Query 37 YSRFFLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRALREVKIRER 96
+ + F E ++G G FGT+ + KL +A+K++ + ++ ++EV+
Sbjct 297 FRQNFTEISRLGSGGFGTVFKAKSKLANKY---YAIKRVKLHSNK-----CIKEVEALAH 348
Query 97 LSHPNIVDYKHSW 109
L HPNIV Y HSW
Sbjct 349 LDHPNIVRYHHSW 361
> mmu:26413 Mapk1, 9030612K14Rik, AA407128, AU018647, C78273,
ERK, Erk2, MAPK2, PRKM2, Prkm1, p41mapk, p42mapk; mitogen-activated
protein kinase 1 (EC:2.7.11.24); K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=358
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 33/57 (57%), Gaps = 3/57 (5%)
Query 47 IGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRALREVKIRERLSHPNIV 103
IG GA+G +C L+++ A+KK+ + + + QR LRE+KI R H NI+
Sbjct 29 IGEGAYGMVCSAYDNLNKV---RVAIKKISPFEHQTYCQRTLREIKILLRFRHENII 82
> bbo:BBOV_I001950 19.m02272; protein kinase domain containing
protein
Length=580
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 40/67 (59%), Gaps = 4/67 (5%)
Query 39 RFFLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWL-QRALREVKIRERL 97
R F++ Q+G+GA+G + L E +++ A+KKL + +DR + A+RE+ + L
Sbjct 191 RNFVKVHQVGQGAYGDVWLAEDIVNQ---SPVALKKLKLNEDREGFPKSAIREIVLLNTL 247
Query 98 SHPNIVD 104
H NIV+
Sbjct 248 KHENIVN 254
> cel:Y39G10AR.3 nekl-1; NEK (NEver in mitosis Kinase) Like family
member (nekl-1); K08857 NIMA (never in mitosis gene a)-related
kinase [EC:2.7.11.1]
Length=998
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 31/105 (29%), Positives = 50/105 (47%), Gaps = 11/105 (10%)
Query 11 HRQRRLQV---EETADYEPIPREFLVTGYYSRFFLERQQIGRGAFGTICLCEHKLDELLL 67
H +R +++ E P PR Y R + +G+GAFG+ L + D L+
Sbjct 142 HNRRIVEIYINNEIMITNPNPRATSAQNRYERI----RTVGKGAFGSAVLYRRREDSSLV 197
Query 68 GEFAVKKLPVGD-DRHWLQRALREVKIRERLSHPNIVDYKHSWLE 111
+K++ + D D + AL EV + R+ HPNI+ Y S+ E
Sbjct 198 ---IIKEINMYDLDSSQRRLALNEVSLLSRIEHPNIIAYYDSFEE 239
> dre:393987 nek4, MGC64176, zgc:64176; NIMA (never in mitosis
gene a)-related kinase 4 (EC:2.7.11.1); K08857 NIMA (never
in mitosis gene a)-related kinase [EC:2.7.11.1]
Length=849
Score = 41.6 bits (96), Expect = 7e-04, Method: Composition-based stats.
Identities = 22/64 (34%), Positives = 37/64 (57%), Gaps = 4/64 (6%)
Query 47 IGRGAFGTICLCEHKLDELLLGEFAVKKLPV-GDDRHWLQRALREVKIRERLSHPNIVDY 105
+G+G++G + L HK D ++ +KKL + R + A +E ++ +L HPNIV Y
Sbjct 10 VGKGSYGEVNLVRHKSDR---KQYVIKKLNLRTSSRRERRAAEQEAQLLSQLKHPNIVMY 66
Query 106 KHSW 109
+ SW
Sbjct 67 RESW 70
> xla:397785 mapk1-b, Xp42, erk, erk2, mapk, mapk1b, mpk1; mitogen-activated
protein kinase 1 (EC:2.7.11.24); K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=361
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 33/57 (57%), Gaps = 3/57 (5%)
Query 47 IGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRALREVKIRERLSHPNIV 103
IG GA+G +C ++++ A+KK+ + + + QR LRE+KI R H NI+
Sbjct 34 IGEGAYGMVCSAHCNINKV---RVAIKKISPFEHQTYCQRTLREIKILLRFKHENII 87
> mmu:330177 Taok3, 2900006A08Rik, A130052D22, A430105I05Rik;
TAO kinase 3 (EC:2.7.11.1); K04429 thousand and one amino acid
protein kinase [EC:2.7.11.1]
Length=898
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 44/78 (56%), Gaps = 9/78 (11%)
Query 40 FFLERQQIGRGAFGTICLCEHK-LDELLLGEFAVKKLP-VGDDRH--WLQRALREVKIRE 95
F++ +IG G+FG + + +E++ AVKK+ G H W Q L+EVK +
Sbjct 23 LFIDLHEIGHGSFGAVYFATNAHTNEVV----AVKKMSYSGKQTHEKW-QDILKEVKFLQ 77
Query 96 RLSHPNIVDYKHSWLEMH 113
+L HPN ++YK +L+ H
Sbjct 78 QLKHPNTIEYKGCYLKEH 95
> xla:398049 wee1-a, MGC81696, wee1a, xe-wee1a, xwee1; wee1 homolog
(EC:2.7.10.2); K06632 wee1-like protein kinase [EC:2.7.11.1]
Length=555
Score = 41.2 bits (95), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 30/84 (35%), Positives = 39/84 (46%), Gaps = 3/84 (3%)
Query 29 REFLVTGYYSRFFLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRAL 88
RE + Y FLE ++IG G FG++ C +LD KK G L AL
Sbjct 198 RETNMGSRYKTEFLEIEKIGAGEFGSVFKCIKRLDGCFYAIKRSKKPLAGSTDEQL--AL 255
Query 89 REVKIRERLS-HPNIVDYKHSWLE 111
REV L HP++V Y +W E
Sbjct 256 REVYAHAVLGHHPHVVRYYSAWAE 279
> ath:AT3G04810 ATNEK2; ATNEK2 (NIMA-RELATED KINASE 2); ATP binding
/ kinase/ protein kinase/ protein serine/threonine kinase;
K08857 NIMA (never in mitosis gene a)-related kinase [EC:2.7.11.1]
Length=606
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 23/68 (33%), Positives = 43/68 (63%), Gaps = 4/68 (5%)
Query 45 QQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQR-ALREVKIRERLSHPNIV 103
+QIG+G+FG+ L HK ++ L + +KK+ + +R A +E+++ ++ +P IV
Sbjct 8 EQIGKGSFGSALLVRHKHEKKL---YVLKKIRLARQTGRTRRSAHQEMELISKIHNPFIV 64
Query 104 DYKHSWLE 111
+YK SW+E
Sbjct 65 EYKDSWVE 72
> ath:AT1G07880 ATMPK13; MAP kinase/ kinase; K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=363
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 37/70 (52%), Gaps = 4/70 (5%)
Query 38 SRFFLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVG-DDRHWLQRALREVKIRER 96
S++ + IGRGA+G +C + E A+KK+ D+R +R LRE+K+
Sbjct 30 SKYIPPIEPIGRGAYGIVCCATNSETN---EEVAIKKIANAFDNRVDAKRTLREIKLLSH 86
Query 97 LSHPNIVDYK 106
+ H N++ K
Sbjct 87 MDHDNVIKIK 96
> hsa:5594 MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2,
p38, p40, p41, p41mapk; mitogen-activated protein kinase
1 (EC:2.7.11.24); K04371 extracellular signal-regulated kinase
1/2 [EC:2.7.11.24]
Length=360
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 33/57 (57%), Gaps = 3/57 (5%)
Query 47 IGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQRALREVKIRERLSHPNIV 103
IG GA+G +C ++++ A+KK+ + + + QR LRE+KI R H NI+
Sbjct 31 IGEGAYGMVCSAYDNVNKV---RVAIKKISPFEHQTYCQRTLREIKILLRFRHENII 84
> ath:AT5G28290 ATNEK3; ATNEK3 (NIMA-RELATED KINASE3); ATP binding
/ kinase/ protein kinase/ protein serine/threonine kinase;
K08857 NIMA (never in mitosis gene a)-related kinase [EC:2.7.11.1]
Length=568
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 43/68 (63%), Gaps = 4/68 (5%)
Query 45 QQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWLQR-ALREVKIRERLSHPNIV 103
+QIG+G+FG+ L HK ++ L + +KK+ + +R A +E+++ ++ +P IV
Sbjct 8 EQIGKGSFGSALLVRHKHEKKL---YVLKKIRLARQTGRTRRSAHQEMELISKIRNPFIV 64
Query 104 DYKHSWLE 111
+YK SW+E
Sbjct 65 EYKDSWVE 72
> cel:C04G6.1 mpk-2; MAP Kinase family member (mpk-2)
Length=605
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 23/64 (35%), Positives = 36/64 (56%), Gaps = 4/64 (6%)
Query 41 FLERQQIGRGAFGTICLCEHKLDELLLGEFAVKKLPVGDDRHWL-QRALREVKIRERLSH 99
+L + +G GA+G +C +D + A+KK+P H L +R+LREV+I L H
Sbjct 57 YLAEENVGAGAYGVVC---KAMDTRNKKQVAIKKIPRAFTAHTLAKRSLREVRILRELLH 113
Query 100 PNIV 103
NI+
Sbjct 114 ENII 117
> ath:AT3G45640 ATMPK3; ATMPK3 (ARABIDOPSIS THALIANA MITOGEN-ACTIVATED
PROTEIN KINASE 3); MAP kinase/ kinase/ protein binding
/ protein kinase; K04371 extracellular signal-regulated
kinase 1/2 [EC:2.7.11.24]
Length=370
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/59 (38%), Positives = 36/59 (61%), Gaps = 6/59 (10%)
Query 47 IGRGAFGTIC-LCEHKLDELLLGEFAVKKLPVGDDRHW-LQRALREVKIRERLSHPNIV 103
IGRGA+G +C + + + +EL+ A+KK+ D H +R LRE+K+ L H NI+
Sbjct 44 IGRGAYGIVCSVLDTETNELV----AMKKIANAFDNHMDAKRTLREIKLLRHLDHENII 98
> xla:443637 jak2, MGC114825; Janus kinase 2 (a protein tyrosine
kinase) (EC:2.7.10.2); K04447 Janus kinase 2 [EC:2.7.10.2]
Length=1129
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 41/121 (33%), Positives = 56/121 (46%), Gaps = 20/121 (16%)
Query 5 FNPYLRHRQRRLQVEETADYEPIPREFLVT-------GYYSRF------FLER-----QQ 46
F P R R L T DYE + L+ G+ F F ER QQ
Sbjct 792 FRPSFRAIIRDLNSLFTPDYELLAETDLLQNTRTGAFGFSGGFEDRDPPFEERHLKFLQQ 851
Query 47 IGRGAFGTICLCEHKLDELLLGE-FAVKKLPVGDDRHWLQRALREVKIRERLSHPNIVDY 105
+G+G FG++ LC + + GE AVKKL + +L+ RE++I + L H NIV Y
Sbjct 852 LGKGNFGSVELCRYDPLQDNTGEVVAVKKLQHTTE-EYLRDFEREIEILKSLQHDNIVRY 910
Query 106 K 106
K
Sbjct 911 K 911
> dre:30298 jak2b, JAK2bCA, jak2; Janus kinase 2b (EC:2.7.10.2
2.7.1.112); K04447 Janus kinase 2 [EC:2.7.10.2]
Length=1126
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/63 (39%), Positives = 37/63 (58%), Gaps = 2/63 (3%)
Query 45 QQIGRGAFGTICLCEHKLDELLLGEF-AVKKLPVGDDRHWLQRALREVKIRERLSHPNIV 103
QQ+G+G FG++ +C + + GE AVKKL H ++ RE++I + L H NIV
Sbjct 838 QQLGKGNFGSVEMCRYDPLQDNTGEVVAVKKLQHSTTEH-IRDFEREIEILKSLQHENIV 896
Query 104 DYK 106
YK
Sbjct 897 KYK 899
Lambda K H
0.326 0.144 0.459
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2052546600
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40